Translate this page into:
Penetrance of missense mutation of OR2AT4 and OR9G1/9 gene polymorphism increases risk factor in azoospermic patients

*Corresponding author: Dr. Ajit Kumar Saxena, Professor and Head, Department of Pathology/Lab Medicine, All India Institute of Medical Sciences Patna, Patna, Bihar, India. draksaxena1@rediffmail.com
-
Received: ,
Accepted: ,
How to cite this article: Saxena AK, Tiwari M, Agarwal M, Shalini. Penetrance of missense mutation of OR2AT4 and OR9G1/9 gene polymorphism increases risk factor in azoospermic patients. J Reprod Healthc Med 2023;4:7.
Abstract
Objectives:
In human, olfactory receptor (ORs) gene polymorphism regulate testicular development and physiological functions during spermatogenesis. However, the etiopathology of OR gene in male infertility is still restricted that required further studies. Basic mechanism involves the identification of peripheral signals based on specific chemoreceptors site that are expressed on the surface of mature spermatozoa inside the female reproductive tract. These receptors belong to G-protein couple receptors (GPCR) of superfamily and the roles of ORs gene linked to sperm chemotaxis through chemokinesis. In the present study, we have studied the role of Olfactory receptor family 2 subfamilies AT member 4 (OR2AT4) and Olfactory receptor family 9 subfamily G member 1/9 (OR9G1/9) genes polymorphism in clinically diagnosed azoospermic patients.
Materials and Methods:
Systematically, GTG banded karyotypes were developed using short term lymphocyte cultures, fluorescence in situ hybridization using SRY probe and PCR based microdeletion of Y-chromosome was characterized using STS marker (SY255). Whole exome sequencing (WES) based analysis was performed in azoospermic patients to identify the involvement of de-novo mutation in OR2AT4 and OR9G1/OR9G9 genes mutation followed by bioinformatics tools used for the analysis to identify changes in respective amino acids.
Results:
OR2AT4 showing missense mutation after substitution of nucleotide (G→A) followed by lack of change in amino acid (Gln→Gln), but in OR9G1/G9 gene showing changes of substitution of nucleotide thymidine into adenine (T→A), whereas valine is replaced by glutamine (Val→ Glu), similarly thymidine is substituted by cysteine (T→C), after decode phenylalanine is replaced by leucine (Phe→Leu). Further, 3D protein helical structure was developed with the help of bioinformatics on the basis of amino acids coded protein and compare with mutated amino acid or truncated protein residues with the help of iTASSER server.
Conclusion:
Present study demonstrate the role of OR2AT4 and OR9G1/9 gene polymorphism elucidates the functional role of protamine in ectopic receptors followed by increasing “risk” of developing male infertility that leads the clinical condition defined as non-obstructive azoospermia.
Keywords
OR family 2 subfamilies AT member 4 and ORs family 9 subfamily G member 1/9 genes
G-protein-coupled receptor
Spermatogenesis
Whole-exome sequencing
INTRODUCTION
Human infertility is a highly complex phenomenon and has poor knowledge of chemosensing during fertilization between healthy, mature male, and female cells, i.e., sperm and ovum. The olfactory receptors (ORs) and G-protein-coupled receptors (GPCRs) constitute the largest gene family in the human genome.[1] During fertilization, mature sperm that reach the female reproductive tract becomes hyperactivated, followed by sequential changes of prefusion through capacitation (acrosomal reaction) and exchange of genetic information between sperm and oocyte. The spermatozoa are exposed to various chemical stimuli originating from their internal physical environment of the female reproductive tract (oocyte) and its surrounding cells to activate surface receptors belonging to GPCRs that transduce the signaling processes outside or within the cell.[2] These signaling pathways lead to responses (sperm) such as changes in electrical activity, motion, and other physiologic functions.
Most of ectopically expressed OR function in sperm remains unknown, and only physiological roles of ORs are documented in various cell types.[3] The activation of OR signal transduction pathway plays a pivotal role in the vast majority of cellular signaling processes, the cytosolic Ca2+ and Na+ concentrations inducing membrane depolarization control in both somatic and germ cells.[4,5] Interestingly, several ORs are expressed predominantly in human sperm cells and previously reported chemotaxis by sperm in response to floral scents (e.g., bourgeonal) due to Ca2+ influx mediated by bourgeonal binding to a receptor protein (hOR17-4)-modified flagellar beating, thus directing sperm cell motility.[6-8] The rationale behind this study is to identify cytogenetic locus and their mutation of the OR family 2 subfamilies AT member 4 (OR2AT4) and ORs family 9 subfamily G member 1/9 (OR9G1/9) gene polymorphism. In male infertility, the study was further extended to predict three-dimensional (3D) protein structure after identification of the gene(s) using whole-exome sequencing (WES) and decodes to identify respective amino acids to explore the knowledge of translational events associated with structural and functional linkage in azoospermic cases.
MATERIAL AND METHODS
The present study included clinically diagnosed randomly selected cases (n = 20) of non-obstructive azoospermia (NOA) of age group 28 years–40 years, visiting the Outpatient Department of General Medicine or Urology Department of All India Institute of Medical Sciences (AIIMS), Patna, that was categorized on the basis of semen analysis[9] and was referred to the Cytogenetic and Molecular Laboratory, Department of Pathology/Lab Medicine, AIIMS, Patna, for molecular genetic analysis. The present study was approved by the Institutional Ethics Committee of the Institute. Family history of the infertile couples was recorded to develop a pedigree tree to find the mode of inheritance if possible and cytogenetic analysis was carried out for karyotypic pattern to confirm the chromosomal aberrations, after written consent from the patients. All patients were clinically diagnosed, and genital examination, ultrasonography, and endocrine analyses were performed. The median age of the proband was 35 years (range: 25–45 years). Blood samples (2.0 mL) were collected under sterile conditions for short-term lymphocyte cultures and GTG-banded karyotyping using cytoscan Olympus system (USA) followed by fluorescence in situ hybridization (FISH) analysis with the help of SRY probe to confirm the extra copy of the Y-chromosome by following procedures established in our laboratory. Briefly, FISH analysis was carried out for the study of X and Y chromosomes using CEP X (DXZ1) (Xp11.1-Xq11.1) Spectrum Green/LSI SRY (Yp11.3) Spectrum Orange, in both interphase and metaphase plates obtained from Abbott-Vysis, Inc. (USA). X-chromosome was identified by a green signal and Y was identified with an orange (red) signal, nuclear deoxyribonucleic acid (DNA) was labeled with 4’,6-diamidino-2-phenylindole (DAPI) as counterstain (blue).[10,11]
Microdeletion of Y-chromosome analysis using polymerase chain reaction (PCR)
Genomic DNA was extracted according to the standard procedure from blood samples according to the manufacturer’s recommendations (Promega Kit USA). DNA contents were measured by NanoDrop spectrophotometer (Thermo USA) and stored at –70°C till further analysis. PCR was performed to analyze the microdeletion in the AZF region of the Y chromosome using specific sequences tagged sites (STS markers Sy255) primers, sense: GTT ACA GGA TTC GGC GTG AT and anti-sense: CTC GTC ATG TGC AGC CAC. PCR amplification was performed in 25 µL reaction volume containing 10 × Tris (pH 8.4), 50 mM KCl, 25 mM MgCl2, 2.5 mM dNTP, 10 pM of oligonucleotides primers, 50–100 ng DNA, and 1U Taq DNA polymerase and PCR condition as 95°C - 5’(94° - 30 s, 56°C - 45 s, 72°C - 45 s), ×35 cycles, 72°C - 5 with amplicon of 126 bp.[10,12]
WGS used for the identification of mutations
Genomic DNA from three randomly selected cases of NOA was isolated and purified before initiation of DNA sequencing was performed by Illumina sequencing platform performed (Xcelris Lab Ahmadabad, India), where the sequencing data were analyzed through http://www.ensembl.org/Homosapiens/Gene/). In brief, purified (2–3 µg) DNA samples were fragmented to approximately 200 bp by sonication (Covaris) and used to prepare single- or paired-end libraries using the SPRI works Fragment Library System I for Illumina Genome Analyzer on the SPRI-TE Nucleic Acid Extractor (Beckman Coulter). Exome enrichment was performed using the NimbleGen Sequence Capture 2.1 M Exome Array, EZ Exome Library (Roche NimbleGen), or SureSelect Human All Exon version 2 or 50 Mb libraries (Agilent Technologies) according to the recommended protocols. Sequencing was performed on GAIIx or HiSeq instruments (Illumina). For each run, the average depth was a minimum of ~ 100× coverage to allow for optimal variant calling. Burrows-wheeler aligner’s maximal exact matches (BWA-MEM) and Genome analysis tool kit (GATK) were used as algorithms for aligning genome sequencing reads to the reference genome (GRCh37/hg19) and variant calling, respectively. Paired-end sequence reads were aligned to the human genome (hg19 assembly) using the BWA program. Local realignment around indels was performed using the GATK software. Subsequently, duplicate reads were removed and single nucleotide variants and indels were identified. Variants were annotated with information from Ensembl release 62 using Ensembl Perl Application Program Interface including single nucleotide polymorphism (SNP) Effect Predictor. The mutated region identifies by (http://www.ensembl.org/Homosapiens/Tools/Blast/Alignment) during analyses of candidate gene variants when compared to genome-wide data using external available databases such as the uniport (https://www.uniprot.org/). The data were further analyzed using large-scale exome sequencing projects including the exome variant table in the ensemble database.
Homology modelling
Due to the unavailability of existing structure, the 3D protein structure is constructed from databases using bioinformatic tool (iTASSER) server for protein homology modeling. The template 2KS9A.pdb is used by the server for the prediction of the 3D OR9G1/9 protein model.[13]
Statistical analysis
The chi-square test was applied to find the significant difference between cases and their respective controls.
RESULTS
Clinically, diagnosed cases (n = 20) of infertility (azoospermic) were further divided on the basis of karyotypic structural (chromatid break, dicentric and ring chromosome), while numerical variations show 46, XY/47, XYY (mosaic), and XXY. Figure 1 shows whole chromosome painting to differentiate between homologous and nonhomologous chromosomes with GTG-banded karyotype having 47, XXY chromosome compliments. The frequency of these aberrations varies significantly with respect to controls (P < 0.05). FISH analysis using SRY probe further confirms the extra copy of Y-chromosome in mosaic or having karyotype of 47, XYY as shown in Figure 2a and b. The study was further extended to confirm these cases (n = 8) of azoospermia using STS markers to confirm the microdeletion of Y-chromosome (AZFc) region and more than 12.00% cases show deletion of 186bp as shown in Figure 3 (arrow). Further, curiosity has been developed to identify “new” gene mutations (SNPs) using WES using biological databases. Three randomly selected cases of NOA were included and WES was performed. Mutation in OR2AT4 and OR9G1/9 genes was identified. First, we have done pairwise alignment for the normal OR2AT4 and OR9G1/9 genes with mutated sequences using NCBI-BLAST (https://blast.ncbi.nlm. nih.gov/Blast.cgi) as shown in Figures 4 and 5 and Table 1, showing the ORs gene polymorphism in heterozygous condition and missense mutation with dbSNP ID of the respective gene. The mutational site on the chromosome presented using genome decorator to highlight the mutation region (https://www.ncbi.nlm.nih.gov/genome/tools/gdp).
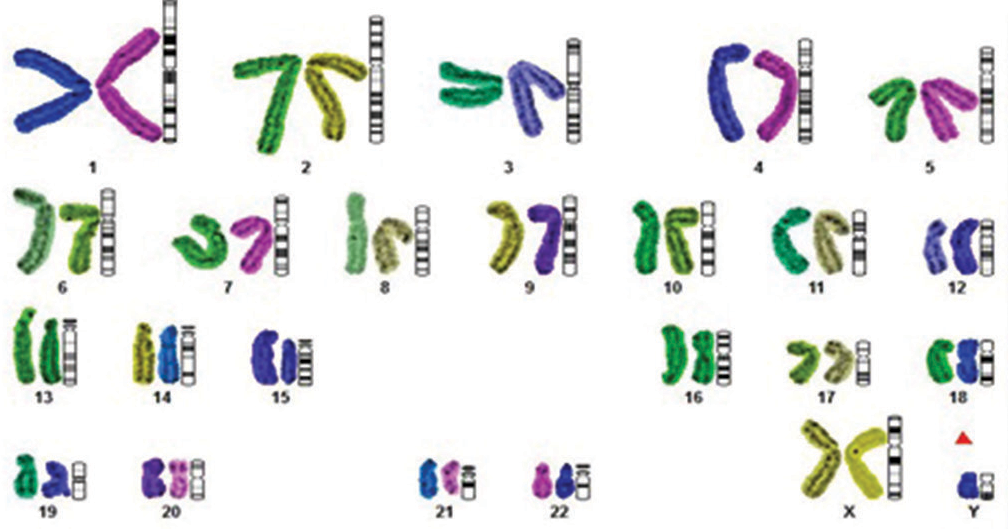
- Representative karyotype (whole chromosome painting) showing the 47, XXY chromosome compliments with differentiating pairing between homozygous and non-homozygous using different color codes with the build software Cytoscan system from Olympus (Japan).
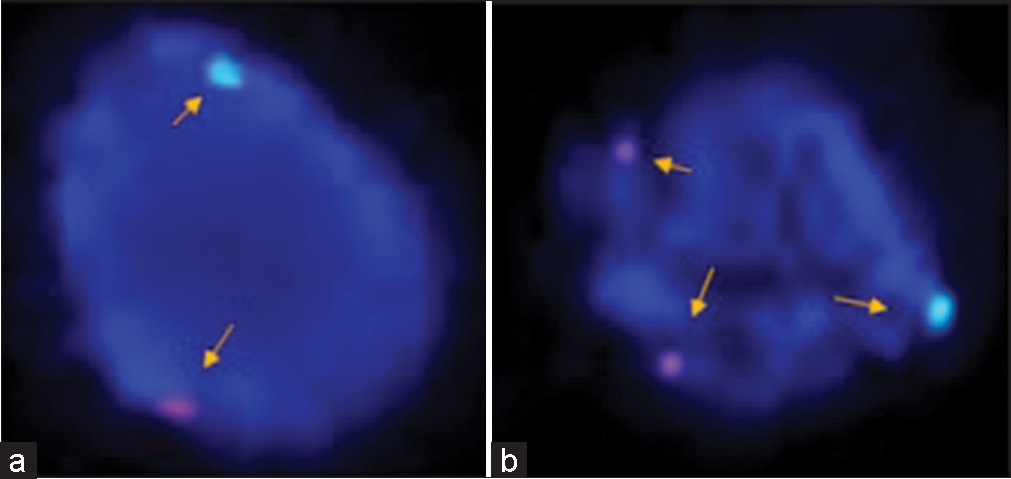
- (a and b) In prophase, fluorescence in situ hybridization (FISH) analysis is showing the identification of sex chromosomes (X and Y) after using specific probes, for the identification of X (green signal) and Y (orange signal) chromosome using Vysis probe (CEP X (DXZ1) (Xp11.1-Xq11.1) Spectrum Green/LSI SRY (Yp11.3) Spectrum Orange, nuclei were counter stained with DAPI (blue). Representative images showing the presence of karyotype XYY in the case of azoospermia. FISH analysis showing X and Y chromosomes (normal a) and an extra copy of the Y-chromosome in prophase (b) showing two red nuclei (arrow) in another case of azoospermia to confirm rare category of Klinefelter’s syndrome.
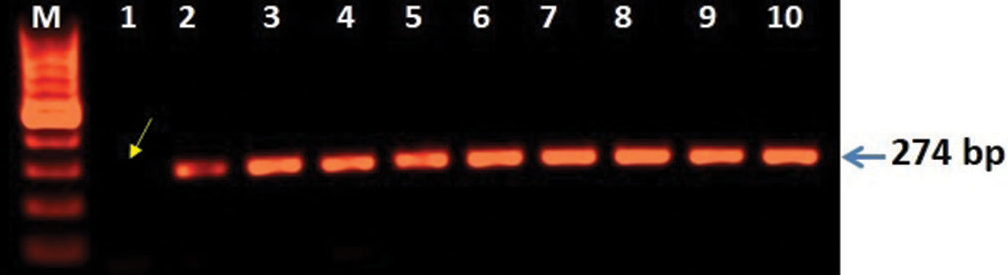
- 3D analysis showing microdeletion of Y-chromosome regions (AZFc). Polymerase chain reaction (PCR)-based analysis was carried out the mutation of SY255 STS marker consist of 126 bp band is completely lost lane-1 (arrow) and lane 2–8 showing lack of mutation when compared with controls (lane 9–10). The amplified PCR products were analyzed on agarose gel (1.5%) electrophoresis and characterized after staining with EtBr using GelDoc system (BioRad USA).
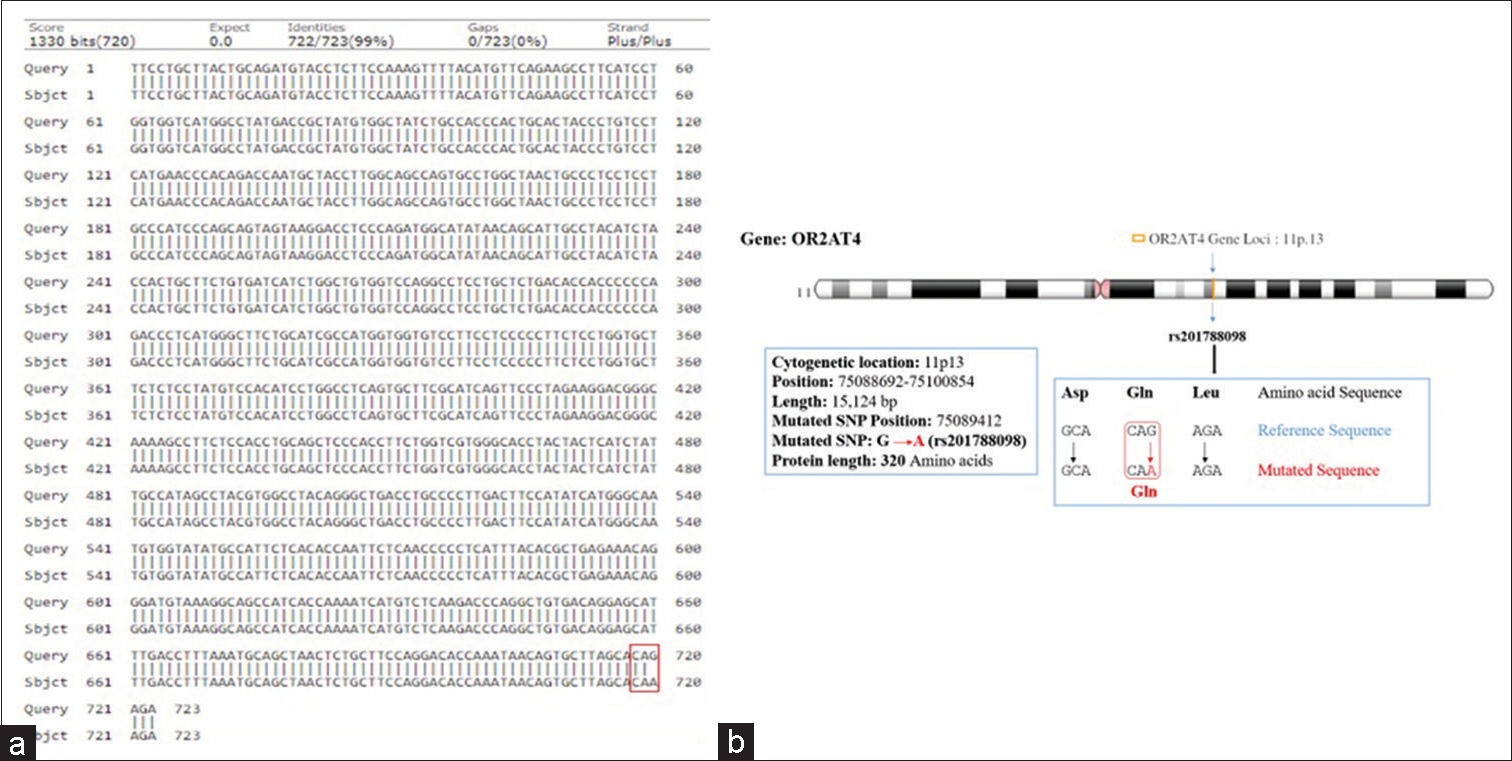
- (a and b) Showing pairwise alignment of normal with mutated sequence (mutated sequence in red square bracket) (a), and chromosome location showing mutational site (locus) of olfactory receptor family 2 subfamilies AT member 4 gene assigned on chromosome-11 (11p.13) as shown in (b).

- (a and b) Showing pairwise alignment of normal and mutated sequence (mutated sequence in red square bracket) (a), and cytogenetic location of mutational locus of olfactory receptor family 9 subfamily G member 1/9gene showing missense mutation assigned on chromosome 11 (11p13) as shown in (b).
| S. No. | Name of gene | dbSNP ID | SNP analysis | Zygosity | Remark |
|---|---|---|---|---|---|
| 1. | OR2AT4 | rs201788098 | G→A | Heterozygous | Missense |
| 2. | OR9G1 | rs79251113 | T→A | Heterozygous | Missense |
| 3. | OR9G9 | rs79288825 | T→C | Heterozygous | Missense |
OR2AT4: Olfactory receptor family 2 subfamilies AT member 4, OR9G1: OR Olfactory receptor family 9 subfamily G member 1, OR9G9: Olfactory receptor family 9 subfamily G member 9
Hence, the mutated SNP position on chromosome spectra has been confirmed with a variant table using a variant table (restriction site) of the Ensemble database (http://asia.ensembl.org/Homosapiens/Gene/VariationGene/Table). The OR2AT4 gene shows mutation of amino acid changes, namely, guanine (G) into alanine (A) (G→A), whereas glutamine (Gln) shows a lack of change, i.e., remains in glutamine in [Figure 4a and b]. However, OR9G1/9 gene mutation of thymine (T) substituted by adenine (A) (T→A) in case of OR9G1, similarly OR9G9 gene, the nucleotide thymine (T) substituted by cytosine (C) (T→C), followed by change of amino acid valine (Val) into glutamate (Glu) and phenylalanine (Phe) changes into leucine (Leu), respectively, as shown in Figure 5a and b. The study was further extended to predict the 3D helical structure of OR9G1/9 constructed by iTASSER server and visualized by PyMol software[14] as shown in Figures 6a and b. Figure 7 shows the location of genes on different sites of sperm, which regulate the functional activity of sperm during spermatogenesis.
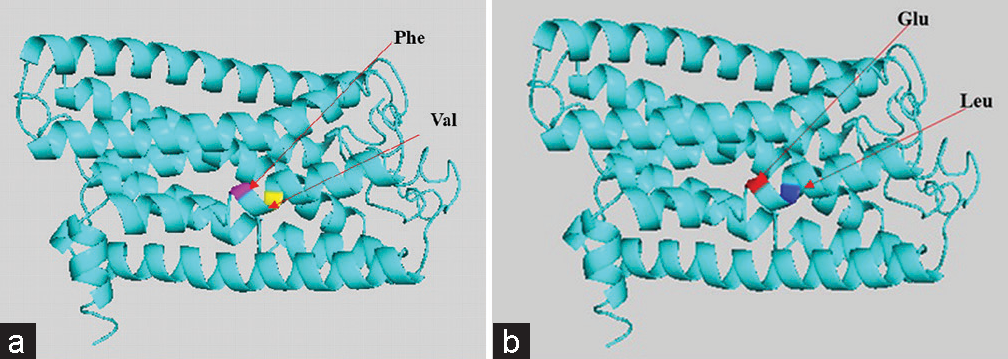
- (a and b) Gene-coded protein 3D helical structure of olfactory receptor family 9 subfamily G member 1/9 showing normal (a) and mutated (b) structure with respected changes of amino acid residues.
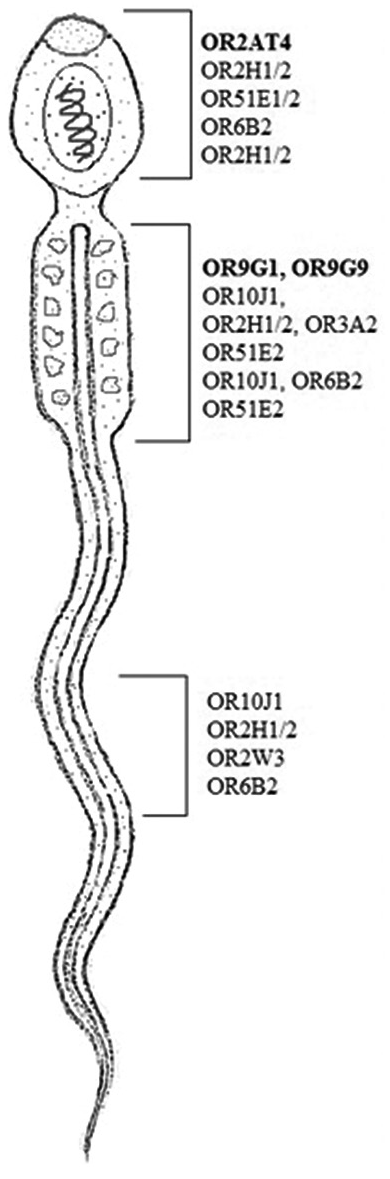
- Olfactory receptor (ORs) gene is located in different sites of human spermatozoa. OR family 2 subfamilies AT member 4 at acrosomic region and ORs family 9 subfamily G member 1/9 found in the middle piece of sperm shown in bold.
DISCUSSION
OR is a chemoreceptor activation protein and is responsible for maintaining the link between the activation of the receptor and sperm behavior. Our study is aimed to identify OR2AT4 and OR9G1/9 gene polymorphism to assess the functional activity during reproduction. The actual mechanisms of controlling the spermatogenesis process have a much more complex phenomenon. There are various factors that work simultaneously under different hormonal conditions to regulate spermatogenesis, but genetic factors cannot be ignored such as karyotyping, FISH analysis, and microdeletion of Y-chromosome by STS markers (AZFc) for confirmation of diagnosis.[10] In accordance with previous studies, our findings also demonstrate the role of OR2AT4 and OR9G1/9 gene in regulating the male reproductive phenomena and dysregulation of such pathways might lead to azoospermic condition. There are some aspects that are poorly understood during crossing-over and synapse formation between homologous and non-homologous chromosomes and also further required more emphasis regarding meiotic events. However, nutritional components affect the anatomy of spermatozoa, physiological, and maturation followed by infertility in humans.[15]
The role of ORs in human spermatozoa modulates the physiological activity of spermatogenesis.[16] Previous findings suggest that ORs family genes (i.e., OR2H1/2, OR3A2, OR51E2, OR10J1, and OR6B2) localized to the midpiece of human spermatozoa and can be activated by the odorant bourgeonal OR4D1 and OR7A5 which are expressed in human testis and sperm and they can be activated by naturally occurring ligands (OR4D1: 5a- androst-16-en-3-one; OR7A5: 4-hydroxy-2,5-dimethyl-3(2H)-furanone.[7,17,18] For all three ORs, their activations were linked to a chemokinetic function in human spermatozoa.[8] However, odorants were also suggested to directly affect the calcium channel in human spermatozoa.[19] In the present study, we investigated that OR2AT4 and OR9G1/9 genes were also expressed in the spermatozoa, i.e., confirmed by WES. Further, we observe that the missense mutation in the OR2AT4 gene shows changes in nucleotide bases but there is no change in amino acid residues, whereas in OR9G1/9 gene shows a missense mutation that modulates the functional activity. Sanger sequencing-based further validation of the mutation is under progress after the inclusion of a large number of cases (data not shown).
In case of OR9G1 gene mutation is resulting in change of Phe into Leu; it is important to note that Phe plays a significant role in the development of acrosomal region of sperm formation as well as in midpiece. The Phe stimulates tyrosine phosphorylation of the sperm flagellum which is responsible for sperm motility. From the present study, it is evident that the OR9G1 mutation affects the motility of sperm and also stimulates apoptosis by replacement of Phe into Leu, suggesting significant increases of mitochondrial superoxide,[20] whereas, in the case of Val and Leu oxidation, the acetyl-CoA reacts with oxaloacetate that can be produced citrate, which could stimulate the Krebs-cycle as energetic substrates used by Sertoli cells for the development of germ cells are essential for spermatocytes and spermatids formation during spermatogenesis.[21]
The OR9G9 gene mutation, showing Val change into glutamate, is an organic molecule that involves in the osmoregularity mechanism. The increase in osmolality from testicular to epididymal fluid should induce the uptake of osmolytes by epididymal sperm to counteract cell shrinkage. These compounds could then be utilized by sperm upon ejaculation into a relatively hypotonic environment in the female tract, by mechanisms of regulatory volume decrease resulting in the efflux of osmolytes and cellular water.[22] Interestingly, our laboratory has previously identified AKP3 and PLOD gene mutations with the help of WES and predicted 3D structure after decoding the genes for ligand binding assay with the anticancer drugs in the cases of male infertility.[23]
CONCLUSION
Our data reveal novel gene mutations in the cases of azoospermia. The present study suggests the involvement of OR genes polymorphism in male infertility. For further establishing the role of OR gene polymorphism, the study has to be extended by including more cases of infertility which will not only validate the role of this novel mutation but also confirm the involvement of OR gene as one of the important factors regulating male fertility. Furthermore, translational research finally concludes that some other unknown factors also play relevant role in sensory receptors for pathophysiological conditions.
Acknowledgments
AKS is thankfully acknowledged to the Director, AIIMS, Patna for continuous support and encouragement. AKS also acknowledges financial support from the Department of Biotechnology project (No. BT/PR14671/MED/12/487/2010), Govt. of India, and finally the patients who participate in the study.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Conflicts of interest
There are no conflicts of interest.
Financial support and sponsorship
Nil.
References
- Particulate adenylate cyclase plays a key role in human sperm olfactory receptor-mediated chemotaxis. J Biol Chem. 2004;279:40194-203.
- [CrossRef] [PubMed] [Google Scholar]
- Human sperm chemotaxis: Both the oocyte and its surrounding cumulus cells secrete sperm chemoattractants. Hum Reprod. 2005;20:761-7.
- [CrossRef] [PubMed] [Google Scholar]
- Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012;45:612-22.
- [CrossRef] [PubMed] [Google Scholar]
- The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11-21.
- [CrossRef] [PubMed] [Google Scholar]
- How the olfactory system makes sense of scents. Nature. 2001;413:211-8.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299:2054-8.
- [CrossRef] [PubMed] [Google Scholar]
- Chemosensory Ca2+ dynamics correlate with diverse behavioural phenotypes in human sperm. J Biol Chem. 2011;286:17311-25.
- [CrossRef] [PubMed] [Google Scholar]
- World health organization reference values for human semen characteristics. Hum Reprod Update. 2010;16:231-45.
- [CrossRef] [PubMed] [Google Scholar]
- Microdeletion of the AZFc locus with high frequency of mosaicism 46,XY/47XYY in cases of non-obstructive azoospermia in eastern population of India. Genet Mol Res. 2019;18:GMR18349.
- [CrossRef] [Google Scholar]
- Penetrance of de novo mutation of USP9Y and PCDH11Y gene in AZF regions of non-obstructive a Azoospermic population in India. Int J Curr Res. 2019;11:1373-9.
- [Google Scholar]
- I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174-81.
- [CrossRef] [PubMed] [Google Scholar]
- Pymol: An open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr. 2002;40:82-92.
- [Google Scholar]
- Functions of essential nutrition for high quality spermatogenesis. Adv Biosci Biotechnol. 2011;2:182-97.
- [CrossRef] [Google Scholar]
- Characterization of the olfactory receptors expressed in human spermatozoa. Front Mol Biosci. 2016;2:73.
- [CrossRef] [PubMed] [Google Scholar]
- Novel function of beta-arrestin2 in the nucleus of mature spermatozoa. J Cell Sci. 2006;119:3047-56.
- [CrossRef] [PubMed] [Google Scholar]
- Sperm-activating odorous substances in human follicular fluid and vaginal secretion: Identification by gas chromatography-olfactometry and Ca2+ imaging. Chempluschem. 2013;78:695-702.
- [CrossRef] [PubMed] [Google Scholar]
- The CatSper channel: A polymodal chemosensor in human sperm. EMBO J. 2012;31:1654-65.
- [CrossRef] [PubMed] [Google Scholar]
- Human spermatozoa possess an IL4I1 l-amino acid oxidase with a potential role in sperm function. Reproduction. 2015;149:587-96.
- [CrossRef] [PubMed] [Google Scholar]
- Metabolism of amino acids by cultured rat Sertoli cells. Metabolism. 2005;54:515-21.
- [CrossRef] [PubMed] [Google Scholar]
- Acquisition of volume regulatory response of sperm upon maturation in the epididymis and the role of the cytoplasmic droplet. Microsc Res Tech. 2003;61:28-38.
- [CrossRef] [PubMed] [Google Scholar]
- Prediction of 3D protein structure based on the mutation of AKAP3 and PLOD3 genes in the case of non-obstructive azoospermia. Int J Fertil Steril. 2020;14:102-9.
- [Google Scholar]






