Translate this page into:
Impact of sperm protamine on semen quality and fertility

*Corresponding author: Kamaraj Elango, Theriogenology Laboratory, Southern Regional Station of ICAR - National Dairy Research Institute, Adugodi, Bengaluru, India. kamarajelango@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Elango K, Kumaresan A, Talluri TR, Raval K, Paul N, Peter ES, et al. Impact of sperm protamine on semen quality and fertility. J Reprod Healthc Med 2022;3:5.
Abstract
Protamines are the nuclear proteins essential for chromatin compaction during spermatogenesis. During chromatin compaction, histones are replaced by transition proteins, which are then replaced by protamines. This process is essential for DNA stability. Protamines are rapidly evolved proteins with high evolutionary variation and encompass positively charged amino acids, especially 48% of arginine. Cysteines present in their sequence allow the formation of disulfide bonds between adjacent protamine molecules. Protamine 1 (PRM1), Protamine 2 (PRM2), and Protamine 3 (PRM3) are reported in mammals. Among these, PRM1 and PRM2 were extensively studied. The normal PRM1 and PRM2 ratios in men, stallions, and mice are 1:1, 3:1, and 1:2, respectively. However, in infertile males, the PRM1: PRM2 ratio is altered due to decreased PRM2 expression, which, in turn, is due to incomplete PRM2 precursor processing and zinc deficiency. In bull, ram, and buck, PRM2 mRNA is present but not PRM2 protein. In mice, rats, bulls, and men, the protamine cluster contains an open reading frame called protamine 3 (gene-4 or protamine-3). The proportion of protamine deficient sperm in the sample is indicative of problems in protamination. Recently, omics technologies, RT-qPCR, and gene knockout-based studies also reported the presence of protamine in sperm. All these semen quality and knockout studies envisage that protamines are indispensable for fertility. Henceforth, protamine-like biomolecules also may be evaluated for fertility prediction or markers in addition to the existing structural and functional attributes of sperm.
Keywords
Protamines
Chromatin compaction
Chromomycin A3
Infertility
Histone
INTRODUCTION
Commonly, the semen quality was evaluated merely by assessing sperm phenotypic characteristics and sperm function. However, nowadays, many techniques have emerged to evaluate the molecular details of the sperm. The sperm proteins, especially nuclear proteins like protamine, were studied in the near past concerning male fertility and semen quality. Friedrich Miescher was the first to report protamine in the semen of salmon fish in 1874.[1] Protamine sulfate neutralizes the anticlotting activity of heparin; therefore, it was used as an antidote for heparin toxicity.[2] It was used in gene therapy because it condenses plasmid Deoxyribonucleic acid (DNA). In addition, it can be used to prevent obesity as it deters low-density lipoprotein. Commonly, protamine is extracted from milt (i.e., fish semen) and used along with insulin to increase the half-life of isophane insulin to treat diabetes. Protamine is also used as a carrier for DNA or Ribonucleic acid (RNA) in drug delivery system in nanopharmaceuticals. The protamine-based RNA vaccines were developed for cancer treatment and also against infectious diseases.[3] On the other hand, from semen biology point of view, protamine is essential during spermatogenesis for chromatin compaction. Protamines are rapidly evolved proteins with a high evolutionary variation. Protamine contains positively charged amino acids, especially rich in arginine (48%). Cysteines present in their sequence allow the formation of disulfide bonds between adjacent protamine molecules.[4,5] Protamine 1 (PRM1), Protamine 2 (PRM2), and Protamine 3 (PRM3) are reported in mammals.[6] Among these, PRM1 and PRM2 have been studied extensively. In bull, ram, and buck spermatozoa, PRM2 mRNA is present but not PRM2 protein. In mice, rats, bulls, and men, the protamine cluster contains an open reading frame called PRM3 (gene-4 or protamine-3). PRM3 is misleading, as it lacks arginine clusters instead of being rich in glutamic acid. Gene4/PRM3 will not bind to DNA and should not be called protamine.[4,7]
The elongating spermatids have soluble polyribosomes in their cytoplasm that synthesizes the protamines and they can pack the genome of sperm. In primates, 10–15% of the sperm genome is packed with histones.[8] The g-globin, e-globin, and telomeric DNA were identified as packed in the mature sperm by histones.[9] Protamines begin to appear in elongating spermatids, during the final process of chromatin condensation. In specific stages of spermatids, the transcription and translation of protamines occur. However, the mRNA of protamines was not reported in Leydig or Sertoli cells. Few studies reported the existence of protamines in the fetal brain, placenta, and kidney, but it is not sure whether this protamine expression is due to artifact or biological expression.[10]
The PRM1 is located in the major groove with every DNA helix turn having one PRM1 molecule.[11] PRM2 is larger than PRM1 and shares around 60% sequence similarity with PRM1. PRM1 and PRM2 can bind up to 11 and 15 base pairs of DNA, respectively.[12] The sperm of horse, tapir, zebra, elephant, hare, and rabbit encompasses two variants of PRM2 (i.e., precursor and processed form). Two additional PRM2 variants were reported in stallion – one containing 62 amino acids (St2a) and the other containing 58 amino acids (St2b).[13] However, the PRM2 content was never reported to be more than 80%. Some species expressed only PRM1, but no species expresses only PRM2. This indicates that PRM2 alone cannot condense the DNA properly. As like two molecules of each H2A, H2B, H3, and H4 create histone octamers to form nucleosomes along with 147 bp of DNA, the PRM1 and PRM2 also interrelate among them and attach to DNA. Protamines can make the molecules of DNA pack together as it counteracts the negative charge of phosphodiester of DNA. The continuation of this process during epididymal transport of spermatozoa can form a disulfide bridge between PRM1 and PRM2 as well as between two molecules of PRM1 that can culminate in condensation of sperm nucleus up to 20 times more than that of the nucleus of somatic cell.[14] This chromatin hypercondensation contributes to the shape of the sperm head. The cleaved PRM2 rather than mature PRM2 was reported to be associated with elongated sperm head in rodents. The arginine content of PRM1 has a role in head shape as it was negatively related to the shape of sperm head in eutherian mammals.[4,10]
FUNCTIONS OF PROTAMINES
The major functions of the PRMs can be enlisted as follows:
These PRMs aid in maintaining the compact and hydrodynamic nucleus with a condensed genome; this is vital because sperm with a hydrodynamic nucleus moves faster
Protects paternal genetic information from free radicals, nucleases, and other mutagens
Removes transcription factors and other proteins, resulting in a blank paternal genetic message, devoid of epigenetic information, therefore allowing its reprogramming by the oocyte
Imprinting of the paternal genome during spermatogenesis
Confer an epigenetic mark on some regions of the sperm genome, affecting its reactivation on fertilization
ROLE OF PROTAMINE IN CHROMATIN COMPACTION
Spermatogenesis involves numerous intricate processes. One of those is chromatin compaction, which happens during spermiogenesis. The process of chromatin compaction is depicted in [Figure 1]. During this process, histones are replaced by transition proteins, which are later replaced by protamines. This process is essential for DNA stability.[10,15] Protamines are the major nuclear proteins in the male germ cells of many species. During spermatogenesis, nucleohistones are replaced by nucleoprotamines in the following steps:
Testis-specific histone variants (TH2B) are incorporated into the sperm DNA
Hyperacetylation reduces the DNA binding affinity of histones and results in DNA relaxation through topoisomerase activation
Replacement of histones with transition proteins 1 and 2 (TNP1 and TNP2)
TNPs are then replaced by protamines to form compact chromatin.[10,14]

- Histone to protamine transition and chromatin compaction.
The histone acetylation (mainly H4 acetylation), phosphorylation, and ubiquitination are responsible for the histone displacement. The chaperons are also required for chromatin remodeling as they displace the histones that were modified post-translationally. The neutralization of the basic proteins, i.e., transition proteins and protamine is vital for chromatin compaction that is achieved by phosphorylation of these proteins.[16]
One or more phosphorylation sites were typically found on the amino-terminal peptide sequence flanking the DNA binding domain and the DNA binding domain of PRM1 in humans, bulls, and stallions. These sites in PRM1 are phosphorylated instantly after its synthesis and also during the entrance of sperm into the oocyte. The major phosphorylation sites are threonine, serine, and tyrosine.[10,17] Another post-translational modification in protamine is the formation of disulfide bonds during the final stages of sperm maturation and epididymal transport, but these disulfide bonds are reduced when sperm enters the oocyte.[14]
METHODS TO EVALUATE CHROMATIN CONDENSATION/PROTAMINE DEFICIENCY IN SPERMATOZOA
To detect the presence and quantify the protamine, currently, the following methods are being adopted:
Toluidine blue staining
Feulgen reaction
Aniline blue staining
Real-time PCR (qPCR)
Transcriptomics (Microarray or next-generation sequencing)
Western blotting
Proteomics
Chromomycin A3 staining
Sperm Chromatin Structure Assay (SCSA).
The toluidine blue staining is performed usually after acid treatment, which stains normal spermatozoa with different shades of green while the sperm with abnormal chromatin stain dark blue to violet.[18] The Schiff ’s reagent is used in Feulgen staining, the normal sperm stain uniform magenta to deep purple color, whereas the sperm with finely granular, coarse clumped, total clearing, and apical clearing appearance are considered to have chromatin alteration.[19] The aniline blue stains sperm with immature nuclei, whereas the sperm that does not stain, is considered mature sperm with well-condensed chromatin.[20] These three methods are indirectly indicating protamine deficiency as abnormal chromatin condensation is usually due to problems in protamination. The qPCR and transcriptomics can be used to estimate protamine mRNA, whereas western blotting and proteomics are for protamine protein estimation, but these techniques are expensive, time-consuming, and require more skill. Therefore, among all these techniques, CMA3 and SCSA are commonly used as they are not consuming much time. CMA3 is a fluorophore that competes with the binding sites of protamines in DNA.[21] The CMA3 binding in sperm indicates protamine deficiency. The sperm stained with CMA3 (protamine deficient sperm) glows with brighter yellow fluorescence, compared to the remaining sperm. The fluorescent microscopic image of CMA3 staining is shown in [Figure 2]. The presence of a higher proportion of CMA3-stained sperm is indicative of a higher proportion of immature sperm in the given ejaculate that can hamper fertility. Protamine reversibly binds with G-C base pairs in the minor groove of the DNA strand. It needs the presence of Mg2+ to bind to the DNA, so the samples are usually treated with a CMA3 solution containing MgCl2 (McIlvaine’s buffer).[22] In SCSA, and Acridine Orange (AO) stain is used to detect damaged DNA by staining intact DNA (dsDNA) as green and denatured (ssDNA) as red. However, SCSA can also be used for detecting protamine deficiency by assessing high DNA stainability (HDS%).[23]

- Fluorescent microscopic representation of chromomycin A3 staining.
ROLE OF PROTAMINE IN SEMEN QUALITY
The protamine content in the fertile patients was equal to protamine in an infertile patient with normal seminal parameters. However, protamine in the fertile patient is not equal to protamine in an infertile patient with abnormal seminal parameters.[24] Sperm with a round head contains more histone and less protamine.[25] The primary and secondary sperm defects positively correlated with chromatin alteration.[19] Protamine levels correlated with viability and DNA integrity.[26]
Protamine ratio
Normal human sperm encompasses PRM1 and PRM2 in equal quantities, but in infertile patients, they are present in altered amounts. Therefore, infertile patients have aberrant PRM1 and PRM2 ratios.[27,70-73] The proportion of PRM1 and PRM2 changes between different species due to variations in PRM2 content. The normal PRM1 and PRM2 ratios in men, stallions, and mice were 1:1, 3:1, and 1:2, respectively. Altered PRM1 and PRM2 ratios indicate DNA fragmentation and infertility. The protamine ratio in fertile and infertile conditions reported in previous studies is depicted in [Table 1]. The altered ratio is due to decreased PRM2 expression in infertile patients, which is due to incomplete PRM2 precursor processing and Zinc deficiency. However, the PRM3:PRM1 ratio remained unaltered in infertile patients.[28,29] An increased proportion of H2B to protamine was reported in infertile men.[30] Khara et al., in 1997,[31] reported a negative correlation (−0.6119) between PRM1/ PRM2 ratio and the IVF rate.
| Fertile men | Infertile men |
|---|---|
| 0.98±0.12 | 1.58±0.24[70] |
| 0.99±0.06 | 1.50±0.05[71] |
| 1.10±0.08 | 3.00±2.84[72] |
| 0.55 to 1.29 range | Outside 0.55–1.29 range[31] |
| 1.01±0.15 | 1.51±0.48 (Oligozoospermia)[73] |
| 1.23±0.65 (Asthenozoospermia)[73] | |
| 1.06±0.01 | <0.8 (13.6%)[15] |
| 0.8–1.2 (46.7%)[15] | |
| >1.2 (39.7%)[15] | |
| 1.00±0.01 | <0.8 (61 patients)[26] |
| 0.8–1.2 (71 patients)[26] | |
| >1.2 (34 patients)[26] |
Relation of sperm protamine with semen quality and male fertility
Various authors have correlated the deficiency in protamines with fertility [Figure 3]. Protamine deficient (chromomycin positive) spermatozoa of buffalo bull (−0.37[32]) have a negative correlation with fertility. Moreover, a higher proportion of protamine deficient sperm was seen in medium and low fertile bulls. Protamine-deficient sperm of human semen (−0.641–−0.40[33-36]) was reported to have a negative correlation with fertility. Likewise, spermatozoa having excessive histones are also negatively correlated (Bull = −0.9[37]) with fertility. Dogan et al. (2015)[38] evaluated the PRM1 content by western blotting, which has a correlation of 0.254 with fertility [Table 2]. These studies collectively indicate that sperm with deficient protamine or excessive histones has less fertilizing potential. In addition, sperm protamine concentrations were negatively correlated with DNA fragmentation.[26] This indicates the role of protamines in protecting sperm DNA from damage. Therefore, sperm with protamine deficiency is prone to DNA damage and subsequent reduction in fertility. Cutoff values of HDS% detected from SCSA range from 10% to 15% in humans and 3.5% in bulls [Table 3].[74-79] If it increases more than these values, it could compromise fertility. Therefore, SCSA will be the best tool for simultaneous assessment of intactness of both DNA and nuclear protein in the chromatin.
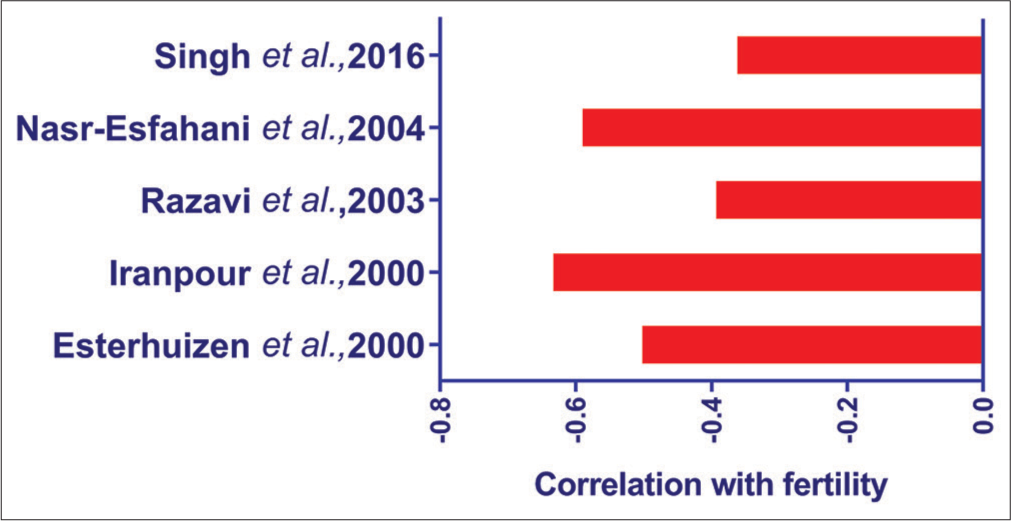
- Proportion of protamine deficient (CMA3 stained) sperm assessed by different authors and its correlation with fertility.
| Parameters | Correlation with fertility | Fertility calculated as | Species |
|---|---|---|---|
| Chromatin maturity (Reduced protamination) Toluidine blue% | −0.61 (P<0.001) | SD of CR | Bull[38] |
| PRM1 content (Western blotting) | 0.254 (P=0.050) | SD of CR | Bull[38] |
| PRM2/PRM1 mRNA ratio (qPCR) | 0.274 (P<0.05) | 28NRR | Stallion[28] |
| PRM1/PRM2 ratio (Protamine1/Protamine2) | −0.6119 (P<0.05) | FR | Human[31] |
| Chromomycin (CMA3) (Protamine deficiency) | −0.37 (P=0.056) | CR | Buffalo bull[32] |
| −0.641 (P<0.001) | FR | Human[33] | |
| −0.401 (P=0.002) | FO-ICSI | Human[35] | |
| −0.598 (P=0.000) | FO-ICSI | Human[36] | |
| −0.51 (P=0.0001) | FR | Human[34] | |
| Aniline blue (Excess Histones) | 0.186 (P=0.174) | FO-ICSI | Human[35] |
| −0.90 (P<0.001) | SD of CR | Bull[37] |
| HDS% | Fertility calculated as | Species |
|---|---|---|
| 15 | PL after IVF ICSI | Human[74] |
| 15 | CP PL after IVF | Human[75] |
| 15 | BP CP IR after IVF ICSI | Human[76] |
| 10 | BP CP after IVF ICSI | Human[77] |
| 15 | CP after IVF ICSI | Human[78] |
| 3.5 | Pregnancy rate 90 d after AI | Bull[79] |
BP: Biochemical pregnancy, CP: Clinical pregnancy, PL: Pregnancy loss, SCD: Sperm chromatin dispersion test, ICSI: Intracytoplasmic sperm injection, HDS: High DNA stainability
PRM1 is the most abundant transcript present in the bovine spermatozoa as per the RNA sequencing results of two studies.[39,40] Protamine mRNA is reported in several species such as human,[41-43] bovine,[32,39,44,45] equine,[46] and porcine[47,48] using various techniques such as RNA-Seq,[32,39,40,46] microarray,[44] and RT-PCR.[39,48] PRM1 is a nuclear protein formed during the condensation of sperm chromatin[4,49] and involved in the process of cell differentiation, chromosome condensation, multicellular organism development, and spermatogenesis. It also has functions such as packaging and stabilizing sperm DNA[44] and the formation of sperm chromatin structure.[50] PRM1 is reported to be abundant in spermatozoa and delivered to the oocyte.[51]
PRM1 is indicative of sperm fertility and is studied based on different fertility indices [Table 4].[80-83] Its concentration was found to be higher in high fertile bulls than in low fertile bulls (P < 0.05) by microarray and RT-PCR analysis.[44] Similarly, bulls in a good quality semen-producing group have shown a higher level of PRM1 expression compared to the poor quality semen-producing group (P < 0.05).[52] In boar, PRM1 was significantly (P < 0.05) found to be higher in the high embryo cleavage group semen samples (n = 10) than in the low embryo cleavage group semen samples (n = 10) by RTPCR.[53] On validating the 27 transcripts observed in RNA-Seq on nine bulls with conception rate scores of −6.7 to 3.6, PRM1 was found to be the only transcript present in all the sires by RT-PCR.[39] Furthermore, on validation of spermatozoal transcripts, PRM1 showed significant (P < 0.05) relation to good quality semen and poor-quality semen of crossbred bulls by RT-PCR.[54] It is said that during cryopreservation, the PRM1 will be affected invariably; however, Mondal et al. (2013)[55] reported that there is no difference in the relative expression of PRM1 between fresh and frozen-thawed epididymal semen from Bos Frontalis by RT-PCR. Using RTPCR, a down-regulated expression of PRM1 was observed in the asthenozoospermic patients.[42] The sperm transcript of the PRM1 gene is present in all livestock species, and its abundance ratio is high irrespective of the semen condition (fresh and frozen) and techniques (RNA-Seq and microarray analysis). It is observed as a full-length coverage transcript having the possibility of protein-coding, and thus, PRM1 could be used as a non-invasive tool for fertility markers.
| Protamine | Inference | Species |
|---|---|---|
| PRM1 mRNA | Low in low fertile | Bull[44] |
| PRM1 mRNA | High in good motile | Bull[52] |
| PRM2 mRNA | Not differ based on motility | |
| PRM1 mRNA | High in low motile sperm | Men[80] |
| PRM1, PRM2 mRNA | Upregulated in bulls with poor motility | Bull[81] |
| PRM1, PRM2 protein | Low in DFI | Men[15] |
| PRM1, PRM2 mRNA | P1, P2 mRNA retention in infertile | Men[15] |
| PRM1, PRM2 protein | Vary in every individual cell, correlate with viability and DNA integrity | Men[26] |
| PRM1, PRM2 mRNA | Decreased in asthenozoospermia | Men[42,82] |
| PRM2 mRNA | Not differ between normal and abnormal spermatogenesis | Men[83] |
| PRM1 mRNA | High in high embryo cleavage group | Boar[53] |
| PRM1 mRNA | No difference in fresh and frozen epididymal sperm | Mithun[55] |
Effect of protamine knockout
Disruption of the coding sequence of one allele in either PRM1 or PRM2 in the embryonic stem cell by homologous recombination and the injection of those embryonic stem cells into blastocyst produced the chimeric mice, which produced the sperm with disturbed PRM1 or PRM2 allele. These sperm had an abnormal nuclear formation, problems in protamine-2 processing, and abnormal sperm function. This shows that both the protamine is indispensable for male fertility and the haploinsufficiency of either PRM1 or PRM2 has resulted in male infertility in mice.[56] Takeda et al.[57] generated the PRM1 lacking female chimeric mice having the oocytes lacking PRM1. These mice produced the male PRM1 (+, −) offspring. The sperm from these male mice were used for in vitro fertilization to produce viable healthy offspring. This study revealed that the sperm from PRM1 (+,−) males can produce healthy offspring, despite having abnormal sperm with high DNA fragmentation, decreased mitochondrial membrane potential, and slight irrationality in expression profile. Schneider et al.[58] used the CRISPR/Cas9 for gene editing in oocytes to produce PRM2 deficient mice. Interestingly, the sperm of heterozygous male mice was having normal motility and head morphology. However, problems in chromatin condensation were noticed in the sperm lacking PRM2 due to the perturbed histone to protamine transition. In addition, these sperm had problems in acrosome formation and membrane abnormalities. This study indicated that in PRM2 deficient mice, heterozygous males were fertile and homozygous males were sterile. TNP1 and TNP2 knockout mice spermatids showed the normal shape of the nucleus, translation suppression, histone disappearance, and protamine deposition but irregular chromatin condensation, PRM2 processing, and DNA breaks. However, the integrity of the genome was preserved in mature spermatids as the intracytoplasmic sperm injection resulted in successful fertilization and produced offspring. Nevertheless, the retention of spermatids in the testis was observed, which resulted in the abnormal and reduced number of sperm in epididymis.[59]
PROTAMINE AND ASSISTED REPRODUCTIVE TECHNOLOGIES (ART) OUTCOME
The interventions in reproduction through ART are emerging continuously. Intracytoplasmic sperm injection (ICSI) is an ART, in which a single spermatozoon is inserted into the oocyte for fertilization. The fertilization capacity was higher in both ICSI and IVF when the sperm with normal levels of protamine mRNA was used.[60] The chromatin maturity measured by Toluidine blue and CMA3 staining was deranged in the patients with poor ART outcomes in terms of fertilization rate.[61] The rate of fertilization and the embryo quality was less in the sperm with high protamination.[62] The histone to protamine ratio (HPR) was negatively correlated with the blastocyst formation rate and delivery rate regardless of the type of ART used.[63] The histone retention and protamine ratio were significantly associated with blastocyst quality after ART.[64]
Mutations in protamine gene and male fertility
The screening for mutation in PRM1 revealed the c.102G > T transversion and c.119G > A, p. Cys40Tyr novel missense mutation in oligospermic individuals. Variants such as c.−107G > C in −15 bp from the transcription initiation site and a c.*51G > C in the 3'UTR of PRM1 were detected in the PRM1 of oligospermic individuals.[65] A total of five SNPs were identified in the Swedish population and seven SNPs were identified in the Spanish population in PRM1 gene. A total of 15 variants were reported in the PRM2 of the Spanish population.[66] The c.65G > A mutation in PRM1 gene and −67C > T mutation in PRM2 promotor region was reported to be associated with spermatogenic failure.[67] The SNP in the PRM2 gene that terminates the translation can cause haploinsufficiency of PRM2 and hence male infertility.[68] A heterozygous SNP in PRM1 that can impair the highly conserved arginine clusters vital for normal DNA binding was also reported in the infertile male patients.[69] All these reports are indicating that the protamine gene without any mutation is essential for male fertility.
CONCLUSION
Protamine is the highly abundant transcript and protein found in spermatozoa, as reported by many studies. Many semen quality-related studies and knockout studies showed that protamine is indispensable for fertility. However, many studies are conducted on humans only. Therefore, the role of protamine, its correlation with fertility, and its cutoff value in domestic animals need to be elucidated by studies in the future. Like protamine, other biomolecules associated with fertility also may be ascertained in the future. In addition to the structural and functional attributes of sperm, protamine-like biomolecules also can be evaluated for fertility prediction and may be used as potential markers for fertility.
Authors’ contributions
KE, AK, and TRT conceptualized the review. All authors were involved in the literature review and development of the manuscript and approved the manuscript for publication.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Die Spermaozoen Einiger Wirbeltiere In: Ein Beitrag zur Histochemie. Basel: Verhandlungen der Naturforschenden Gesellschaft in Basel VI (in German); 1874. p. :138-208.
- [Google Scholar]
- Precipitation and neutralization of heparin from different sources by protamine sulfate. Pharmaceuticals. 2017;10:59.
- [CrossRef] [PubMed] [Google Scholar]
- Use of protamine in nanopharmaceuticals A review. Nanomaterials. 2021;11:1508.
- [CrossRef] [PubMed] [Google Scholar]
- Protamines and male infertility. Hum Reprod Update. 2006;12:417-35.
- [CrossRef] [PubMed] [Google Scholar]
- Aberrant protamine content in sperm and consequential implications for infertility treatment. Hum Fertil. 2014;17:80-9.
- [CrossRef] [PubMed] [Google Scholar]
- Protamine 3 expressions in crossbred bull spermatozoa may not be a prognostic marker for differentiating good and poor quality semen. Afr J Biotechnol. 2014;13:13535.
- [CrossRef] [Google Scholar]
- Sperm nuclear protamines: A checkpoint to control sperm chromatin quality. Anat Histol Embryol. 2018;47:273-9.
- [CrossRef] [PubMed] [Google Scholar]
- Telomere dynamics throughout spermatogenesis. Genes. 2019;10:525.
- [CrossRef] [PubMed] [Google Scholar]
- Chromatin structure of telomere domain in human sperm. Biochem Biophys Res Commun. 2000;279:213-8.
- [CrossRef] [PubMed] [Google Scholar]
- The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:1-8.
- [CrossRef] [PubMed] [Google Scholar]
- Nuclear organization of the protamine locus. Soc Reprod Fertil Suppl. 2007;64:1-12.
- [CrossRef] [PubMed] [Google Scholar]
- Zinc is sufficiently abundant within mammalian sperm nuclei to bind stoichiometrically with protamine 2. Mol Reprod Dev. 2000;56:512-9.
- [CrossRef] [Google Scholar]
- Comparison of partial amino acid sequences of two protamine 2 variants from stallion sperm structural evidence that the variants are products of different genes. Febs Lett. 1989;244:199-202.
- [CrossRef] [Google Scholar]
- Formation of native-like mammalian sperm cell chromatin with folded bull protamine. J Biol Chem. 2004;279:20088-95.
- [CrossRef] [PubMed] [Google Scholar]
- DNA integrity is compromised in protamine-deficient human sperm. J Androl. 2005;26:741-8.
- [CrossRef] [PubMed] [Google Scholar]
- Packaging paternal chromosomes with protamine. Nat Genet. 2001;28:10-2.
- [CrossRef] [PubMed] [Google Scholar]
- Tyrosine phosphorylation, thiol status, and protein tyrosine phosphatase in rat epididymal spermatozoa. Biol Reprod. 2004;71:1009-15.
- [CrossRef] [PubMed] [Google Scholar]
- Morphometric features and chromatin condensation abnormalities evaluated by toluidine blue staining in bull spermatozoa. J Morphol Sci. 2017;22:85-90.
- [Google Scholar]
- Comparison between the toluidine blue stain and the Feulgen reaction for evaluation of rabbit sperm chromatin condensation and their relationship with sperm morphology. Theriogenology. 2004;62:398-402.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative study of cytochemical tests for sperm chromatin integrity. J Androl. 2001;22:45-53.
- [Google Scholar]
- Use of chromomycin A3 staining in bovine sperm cells for detection of protamine deficiency. Biotech Histochem. 2009;84:79-83.
- [CrossRef] [PubMed] [Google Scholar]
- Chromomycin A3 staining, sperm chromatin structure assay and hyaluronic acid binding assay as predictors for assisted reproductive outcome. Reprod Biomed Online. 2009;19:671-84.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical aspects of sperm DNA fragmentation detection and male infertility. Theriogenology. 2006;65:979-91.
- [CrossRef] [PubMed] [Google Scholar]
- Electrophoretic patterns of spermatozoal nucleoproteins (NP) in fertile men and infertility patients and comparison with NP of somatic cells: Nukleoproteinfraktionen von spermien fertiler männer und infertilitätspatienten im vergleich mit den nukleoproteinen somatischer zellen. Andrologia. 1990;22:217-24.
- [CrossRef] [PubMed] [Google Scholar]
- Anomalous distribution of nuclear basic proteins in round-headed human spermatozoa: Anormale verteilung basischer kern-proteine in menschlichen rundköpfigen spermatozoen. Andrologia. 1990;22:549-55.
- [CrossRef] [PubMed] [Google Scholar]
- Protamine levels vary between individual sperm cells of infertile human males and correlate with viability and DNA integrity. J Androl. 2006;27:890-8.
- [CrossRef] [PubMed] [Google Scholar]
- Protamine alterations in human spermatozoa. Adv Exp Med Biol. 2014;791:83-102.
- [CrossRef] [PubMed] [Google Scholar]
- Protamine mRNA ratio in stallion spermatozoa correlates with mare fecundity. Andrology. 2014;2:521-30.
- [CrossRef] [PubMed] [Google Scholar]
- The impact of sperm protamine deficiency and sperm DNA damage on human male fertility: A systematic review and meta-analysis. Andrology. 2016;4:789-99.
- [CrossRef] [PubMed] [Google Scholar]
- Sperm nuclear histone to protamine ratio in fertile and infertile men: Evidence of heterogeneous subpopulations of spermatozoa in the ejaculate. J Androl. 2006;27:414-20.
- [CrossRef] [PubMed] [Google Scholar]
- Human protamines and male infertility. J Assist Reprod Genet. 1997;14:282-90.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of suitable combinations of in vitro sperm-function test for the prediction of fertility in buffalo bull. Theriogenology. 2016;86:2263-71.
- [CrossRef] [PubMed] [Google Scholar]
- Chromomycin A3 staining as a useful tool for evaluation of male fertility. J Assist Reprod Genet. 2000;17:60-6.
- [CrossRef] [PubMed] [Google Scholar]
- Sperm chromatin packaging as an indicator of in-vitro fertilization rates. Hum Reprod. 2000;15:657-61.
- [CrossRef] [PubMed] [Google Scholar]
- Effect of human sperm chromatin anomalies on fertilization outcome post-ICSI. Andrologia. 2003;35:238-43.
- [CrossRef] [PubMed] [Google Scholar]
- Relationship between protamine deficiency with fertilization rate and incidence of sperm premature chromosomal condensation post-ICSI. Andrologia. 2004;36:95-100.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular morphology and function of bull spermatozoa linked to histones and associated with fertility. Reproduction. 2013;146:263-72.
- [CrossRef] [PubMed] [Google Scholar]
- Sperm protamine-status correlates to the fertility of breeding bulls. Biol Reprod. 2015;92:92.
- [CrossRef] [PubMed] [Google Scholar]
- Cryopreserved bovine spermatozoal transcript profile as revealed by high-throughput ribonucleic acid sequencing. Biol Reprod. 2013;88:49.
- [CrossRef] [PubMed] [Google Scholar]
- Occurrence and functional significance of the transcriptome in bovine (Bos taurus) spermatozoa. Sci Rep. 2017;7:42392.
- [CrossRef] [PubMed] [Google Scholar]
- Differential gene expression in preimplantation embryos from mouse oocytes injected with round spermatids or spermatozoa. Hum Reprod. 2001;16:1449-56.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of protamines 1 and 2 transcript contents in spermatozoa from asthenozoospermic men. Folia Histochem Cytobiol. 2007;45:109-13.
- [Google Scholar]
- Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 2013;41:4104-17.
- [CrossRef] [PubMed] [Google Scholar]
- Transcriptome analysis of bull spermatozoa: Implications for male fertility. Reprod Biomed Online. 2010;21:312-24.
- [CrossRef] [PubMed] [Google Scholar]
- Insight into bovine (Bos indicus) spermatozoal whole transcriptome profile. Theriogenology. 2019;129:8-13.
- [CrossRef] [PubMed] [Google Scholar]
- Stallion sperm transcriptome comprises functionally coherent coding and regulatory RNAs as revealed by microarray analysis and RNA-seq. PLoS One. 2013;8:e56535.
- [CrossRef] [PubMed] [Google Scholar]
- Analysis of selected transcript levels in porcine spermatozoa, oocytes, zygotes and two-cell stage embryos. Reprod Fertil Dev. 2008;20:513-8.
- [CrossRef] [PubMed] [Google Scholar]
- Identification and sequencing of remnant messenger RNAs found in domestic swine (Sus scrofa) fresh ejaculated spermatozoa. Anim Reprod Sci. 2009;113:143-55.
- [CrossRef] [PubMed] [Google Scholar]
- Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog Nucleic Acid Res Mol Biol. 1991;40:25-94.
- [CrossRef] [Google Scholar]
- DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry. 1996;23:263-71.
- [CrossRef] [Google Scholar]
- Current status of sperm functional genomics and its diagnostic potential of fertility in bovine (Bos taurus) Syst Biol Reprod Med. 2018;64:484-501.
- [CrossRef] [PubMed] [Google Scholar]
- Differential expression of protamine 1 and 2 genes in mature spermatozoa of normal and motility impaired semen producing crossbred Frieswal (HF× Sahiwal) bulls. Res Vet Sci. 2013;94:256-62.
- [CrossRef] [PubMed] [Google Scholar]
- Quantitative analysis of sperm mRNA in the pig: Relationship with early embryo development and capacitation. Reprod Fertil Dev. 2013;25:807-17.
- [CrossRef] [PubMed] [Google Scholar]
- Database on spermatozoa transcriptogram of catagorised Frieswal crossbred (Holstein Friesian X Sahiwal) bulls. Theriogenology. 2019;129:130-45.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization and gene expression profiling of epididymal sperm collected from dead mithun (Bos frontalis) bulls and its preservation. Int J Biotechnol Bioeng. 2013;4:535-42.
- [Google Scholar]
- Haploinsufficiency of protamine-1 or-2 causes infertility in mice. Nat Genet. 2001;28:82-6.
- [CrossRef] [PubMed] [Google Scholar]
- Viable offspring obtained from Prm1-deficient sperm in mice. Sci Rep. 2016;6:27409.
- [CrossRef] [PubMed] [Google Scholar]
- Re-visiting the Protamine-2 locus: Deletion, but not haploinsufficiency, renders male mice infertile. Sci Rep. 2016;6:36764.
- [CrossRef] [PubMed] [Google Scholar]
- Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis. 2004;38:200-13.
- [CrossRef] [PubMed] [Google Scholar]
- The sperm protamine mRNA ratio as a clinical parameter to estimate the fertilizing potential of men taking part in an ART programme. Hum Reprod. 2013;28:969-78.
- [CrossRef] [PubMed] [Google Scholar]
- Protamines and DNA integrity as a biomarkers of sperm quality and assisted conception outcome. Andrologia. 2022;54:e14418.
- [CrossRef] [Google Scholar]
- Relationships between human sperm protamines, DNA damage and assisted reproduction outcomes. Reprod Biomed Online. 2011;23:724-34.
- [CrossRef] [PubMed] [Google Scholar]
- The impact of histones linked to sperm chromatin on embryo development and ART outcome. Andrology. 2018;6:436-45.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative analysis of three sperm DNA damage assays and sperm nuclear protein content in couples undergoing assisted reproduction treatment. Hum Reprod. 2014;29:904-17.
- [CrossRef] [PubMed] [Google Scholar]
- Mutations in the protamine 1 gene associated with male infertility. Mol Hum Reprod. 2007;13:461-4.
- [CrossRef] [PubMed] [Google Scholar]
- Polymorphisms, haplotypes and mutations in the protamine 1 and 2 genes. Int J Androl. 2011;34:470-85.
- [CrossRef] [PubMed] [Google Scholar]
- Mutations in the protamine locus: Association with spermatogenic failure? Mol Hum Reprod. 2009;15:733-8.
- [CrossRef] [PubMed] [Google Scholar]
- Single nucleotide polymorphisms in the protamine-1 and-2 genes of fertile and infertile human male populations. Mol Hum Reprod. 2003;9:69-73.
- [CrossRef] [PubMed] [Google Scholar]
- An SNP in protamine 1: A possible genetic cause of male infertility? J Med Genet. 2006;43:382-4.
- [CrossRef] [PubMed] [Google Scholar]
- Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia. 1988;44:52-5.
- [CrossRef] [PubMed] [Google Scholar]
- Human male infertility may be due to a decrease of the protamine P2 content in sperm chromatin. Mol Reprod Dev. 1993;34:53-7.
- [CrossRef] [PubMed] [Google Scholar]
- Complete selective absence of protamine P2 in humans. J Biol Chem. 1993;268:10553-7.
- [CrossRef] [Google Scholar]
- Marked differences in protamine content and P1/P2 ratios in sperm cells from percoll fractions between patients and controls. J Androl. 2003;24:438-47.
- [CrossRef] [PubMed] [Google Scholar]
- Sperm chromatin structure assay parameters are not related to fertilization rates, embryo quality, and pregnancy rates in in vitro fertilization and intracytoplasmic sperm injection, but might be related to spontaneous abortion rates. Fertil Steril. 2008;90:352-9.
- [CrossRef] [PubMed] [Google Scholar]
- Sperm chromatin structure assay results after swim-up are related only to embryo quality but not to fertilization and pregnancy rates following IVF. Asian J Androl. 2011;13:862.
- [CrossRef] [PubMed] [Google Scholar]
- The sperm chromatin structure assay as a diagnostic tool in the human fertility clinic. Hum Reprod. 2006;21:1576-82.
- [CrossRef] [PubMed] [Google Scholar]
- The predictive value of sperm chromatin structure assay (SCSA) parameters for the outcome of intrauterine insemination, IVF and ICSI. Hum Reprod. 2004;19:1401-8.
- [CrossRef] [PubMed] [Google Scholar]
- Relationship between the outcomes of assisted reproductive techniques and sperm DNA fragmentation as measured by the sperm chromatin structure assay. Fertil Steril. 2003;80:895-902.
- [CrossRef] [Google Scholar]
- Bull and boar sperm DNA integrity evaluated by sperm chromatin structure assay in the Czech Republic. Vet Med (Praha). 2004;49:1-8.
- [CrossRef] [Google Scholar]
- Analysis and significance of mRNA in human ejaculated sperm from normozoospermic donors: Relationship to sperm motility and capacitation. Mol Hum Reprod. 2004;10:535-41.
- [CrossRef] [PubMed] [Google Scholar]
- Profiling of sperm gene transcripts in crossbred (Bos taurus x Bos indicus) bulls. Anim Reprod Sci. 2017;177:25-34.
- [CrossRef] [PubMed] [Google Scholar]
- Differential RNAs in the sperm cells of asthenozoospermic patients. Hum Reprod. 2012;27:1431-8.
- [CrossRef] [PubMed] [Google Scholar]
- Decreased protamine-1 transcript levels in testes from infertile men. Mol Hum Reprod. 2003;9:331-6.
- [CrossRef] [PubMed] [Google Scholar]






