Translate this page into:
Omics-based cutting-edge technologies for identifying predictive biomarkers to measure the impact of air borne particulate matter exposure on male reproductive health

*Corresponding author: Pradyumna Kumar Mishra, Division of Environmental Biotechnology, Genetics and Molecular Biology, ICMR-National Institute for Research in Environmental Health, Bhopal, Madhya Pradesh, India. pkm_8bh@yahoo.co.uk
-
Received: ,
Accepted: ,
How to cite this article: Kumari R, Kaur P, Verma SK, Ratre P, Mishra PK. Omics-based cutting-edge technologies for identifying predictive biomarkers to measure the impact of airborne particulate matter exposure on male reproductive health. J Reprod Healthc Med. 2024;5:2. doi: 10.25259/JRHM_25_2023
Abstract
The reproductive lifespan of an individual is a critical determinant of their health, population dynamics, and aging. Research has established a clear association between environmental air pollution, particulate matter (PM), and reproductive health. Recent studies have focused on the impact of air pollution on male reproductive health. Chronic or acute exposure to airborne PM0.1, PM2.5, and PM10 has been found to trigger mitochondrial oxidative stress, double-strand DNA breaks, epigenetic modifications, and endocrine disruption in male reproductive tract functions. Consequently, identifying and validating PM-associated predictive biomarkers, including genes, transcripts, epimutations, proteins, and metabolites, hold promise for improving male reproductive efficiency. Omics-based techniques, such as next-generation sequencing, comparative genomic hybridization, genome-wide association studies, single-cell RNA sequencing, microarray analysis, mass spectroscopy, 2D gel electrophoresis, Raman spectroscopy, near-infrared spectroscopy, and nuclear magnetic resonance, have provided crucial insights into the pathological mechanisms underlying air pollution-related male reproductive health issues. This article presents a comprehensive assessment of existing evidence in this field, offering a methodical examination of findings that hold immense potential for addressing the adverse effects of air pollution on male reproductive health.
Keywords
Air pollution
Circulating epigenetic signatures
Biomarkers
Mitochondrial epigenetics
Human biomonitoring
Environmental health
INTRODUCTION
The widespread exposure to hazardous environmental pollutants significantly threatens human reproduction. Reproductive health care, encompassing all phases of life, is dedicated to safeguarding the intricate processes, functions, and systems involved in reproduction.[1,2] These findings underscore the urgent need for comprehensive measures to mitigate the impact of environmental pollutants on reproductive health and ensure the well-being of future generations. Changes in lifestyle variables are one of the most prominent factors that have contributed to an increase in the prevalence of infertility in recent decades.[3] Particulate matter (PM) has been identified as the most detrimental to human health among the various components of air pollution. PM, encompassing particles of different sizes and compositions, enters the respiratory system through inhalation, contributing to prolonged exposure to ambient air pollution and being associated with detrimental effects on respiratory,[4] cardiovascular diseases,[5] reproductive malfunctions,[6] and cancer.[7] Recent findings indicate that ambient air pollution exposure may also adversely affect male reproductive health and fertility. Individuals who are already unwell, along with children and older people, are particularly vulnerable to the more severe impacts stemming from such pollution exposure. Air pollution stands as one of the foremost global environmental contributors to diseases, impacting nearly 2.4 billion individuals who face perilous levels of indoor pollution. Moreover, a staggering 99% of the world’s population emits air pollutants, surpassing the guidelines set by the World Health Organization. Consequently, the adverse effects of air pollution led to more than 7 million premature deaths annually. PM is a complex mixture comprising acids, sulfates, nitrates, metals, dust and soil particles, and organic molecules. PM’s size and elemental composition have been associated with negative health impacts.[8,9] This pollution source contains biological material, organic chemicals, hydrocarbons, acid aerosols, and metals from anthropogenic and natural sources.[10] Notably, PM from anthropogenic sources tends to contain higher levels of polycyclic aromatic hydrocarbons, semiquinones, metals, and transition metals, contributing to increased toxicity.[11] Neonates, characterized by underdeveloped immune systems and frequent breathing, stand out as one of the most vulnerable subsets of the population to the detrimental impacts of air pollution. Exposure during crucial developmental windows may result in morbidity, manifesting in impaired pulmonary and cardiovascular functioning and potential postnatal mortality.[12] Although the precise effects of neonatal air pollution on reproductive functions are not yet fully understood, existing data indicate that adults, who are comparatively less vulnerable, can develop asthma and cardiovascular problems due to air pollution exposure.[13] Furthermore, adult reproductive processes are affected by air pollution exposure, suggesting that similar alterations may occur in neonates, who are inherently more susceptible to air pollution than adults.[14,15]
MOLECULAR MECHANISMS UNDERLYING PM-INDUCED MALE REPRODUCTIVE TRACT DYSFUNCTIONS
Over the past decade, infertility rates have seen a significant surge, with advancing maternal age recognized as a major contributor. However, both male factors, alongside changes in environmental conditions, may also play crucial roles in this concerning trend. Notably, nitrogen dioxide and PM with a diameter of ten micrometers or less (PM10) emerge as pivotal pollutants negatively impacting fertility and live birth outcomes.[16] Air pollution has been shown to detrimentally affect male gametogenesis, influencing the quantity and quality of gametes. These effects extend to embryonic development, highlighting the pervasive impact of environmental conditions on reproductive health.[3] Several proposed pathways in the literature elucidate the relationship between air pollution and fertility. These include hormonal alterations due to endocrine disruptors, oxidative stress induction, cell DNA alteration, and epigenetic modifications. Air pollutants can influence hormones by activating the aryl hydrocarbon receptor, estrogen, and androgen receptors.[17] In addition, their potential to act as pro-oxidants and free radical producers, leading to oxidative stress and inflammatory responses, is a common biological mechanism for their adverse effects.[18] Certain contaminants can induce epigenetic alterations, such as DNA methylation and histone modifications, potentially impacting future generations.[3] From pre-conception to adulthood, the critical periods of human development are susceptible to environmental toxins, shaping reproductive health throughout life. Both animal and human gametogenesis suffer adverse effects from major air pollutants, resulting in diminished reproductive performance. The key mechanisms through which these pollutants impact include altering the endocrine system, enhancing oxidative stress and inflammation, and activating specific targets capable of stimulating mitogen-activated protein kinase (MAPK) signaling [Figure 1]. Ultrafine particles, characterized by their reduced size and composition featuring organic compounds and metals, have the potential to cause oxygen radical injury and genomic consequences within the male reproductive axis. Earlier investigations substantiated that exposure to ultrafine PM0.1 initiates the impairment of mitochondrial redox homeostasis. This impairment is facilitated through a PI-3-kinase-mediated DNA damage response, ultimately activating a nuclear factor-kappa B (NF-kb) self-cascading inflammatory loop. This loop demonstrates the capacity to influence diverse genomic processes. Reactive oxygen species (ROS) have the capacity to initiate lipid peroxidation by interacting with polyunsaturated lipids on the plasma membrane, thereby facilitating the breakdown of lipids and transmembrane proteins. This process contributes to tissue injury. In addition, ROS can stimulate the release of proinflammatory cytokines, leading to the accumulation of neutrophils and other phagocytic cells that further amplify ROS production, culminating in inflammation and metabolic injuries. The ROS-mediated MAPK signaling pathway has been directly associated with PM2.5 exposure, evident in the pronounced increases in kinase phosphorylation.[19] This disruption, in turn, compromises the integrity of the blood testis barrier (BTB), leading to impaired semen quality. This disruption, in turn, compromises the integrity of the BTB, leading to impaired semen quality. The extensively investigated mechanisms encompass inflammation, apoptosis induction, disruption or harm to the BTB, and oxidative stress. Oxidative stress is characterized by an imbalance between the production of ROS and the activity of scavenging antioxidants.[15] There is a well-established understanding that heightened ROS generation can undermine fertilizing capacity by diminishing sperm count and impairing sperm motility. Moreover, elevated levels of antioxidants, notably superoxide dismutase and catalase, have been demonstrated to impede various sperm functions, creating an imbalance between oxidation and reduction. Normal spermatogenesis relies on the integrity of the BTB, a protective boundary separating testicular seminiferous tubules from blood vessels. Supporting cells, particularly Sertoli cells, not only supply nutrients to spermatogenic cells but also contribute to the formation of the BTB. This barrier serves as a defense against the entry of toxic and harmful substances into the seminiferous tubules, thereby safeguarding the safety and homeostasis of the spermatogenic microenvironment. Connexin43 (Cx43) protein has been identified as the primary component of gap junctions in this context. Deficiency in Cx43 can lead not only to disorders in sperm production but also to abnormal proliferation of Sertoli cells, potentially compromising the integrity of the blood testis barrier and diminishing sperm quality. Moreover, studies have revealed that PM2.5 exposure induces excessive ROS production within the testis through the phosphatidylinositol 3-kinase (PI3K) and protein kinase B (Akt) signaling pathways. Elevated production is further promoted by phosphorylated Akt, ultimately leading to oxidative stress impairment through the PI3K/Akt signaling pathway and disrupting spermatogenesis in male rats. Furthermore, the overproduction of ROS has the potential to cause DNA damage by modifying or breaking DNA strands. These lesions may result in base substitutions, insertions, deletions, or rearrangements, thereby affecting the coding or regulatory regions of genes. Animal models examining the effects of pollutants on embryo development and reproductive success underscore the broader implications of air pollution on reproduction [Figure 2].[20] Environmental toxins are considered potential causal agents contributing to the rising infertility rates, given their ubiquitous presence in the environment and almost unavoidable human exposure. Chemical pollutants, including those from occupational exposure and animal trials, have negatively impacted fertility.[21,22] Widespread exposure to environmental toxins has been identified as a serious health concern, particularly in disrupting hormones critical for healthy human reproduction and development.[23] Understanding the molecular associations between gene-environment interactions and human reproduction poses significant methodological challenges. The epigenomic landscape is recognized as highly relevant to reproductive health, playing a substantial role in the complex interplay of genotype-phenotype relationships.[24] While challenges persist, particularly in critical vulnerability windows from conception to 37–40 weeks of gestation, novel methodologies are essential for comprehending the intricacies of human reproduction and improving population health.[25,26] Recent breakthroughs in high-throughput techniques such as genomics, transcriptomics, proteomics, and metabolomics offer promising avenues for enhanced diagnoses and therapies for male infertility.
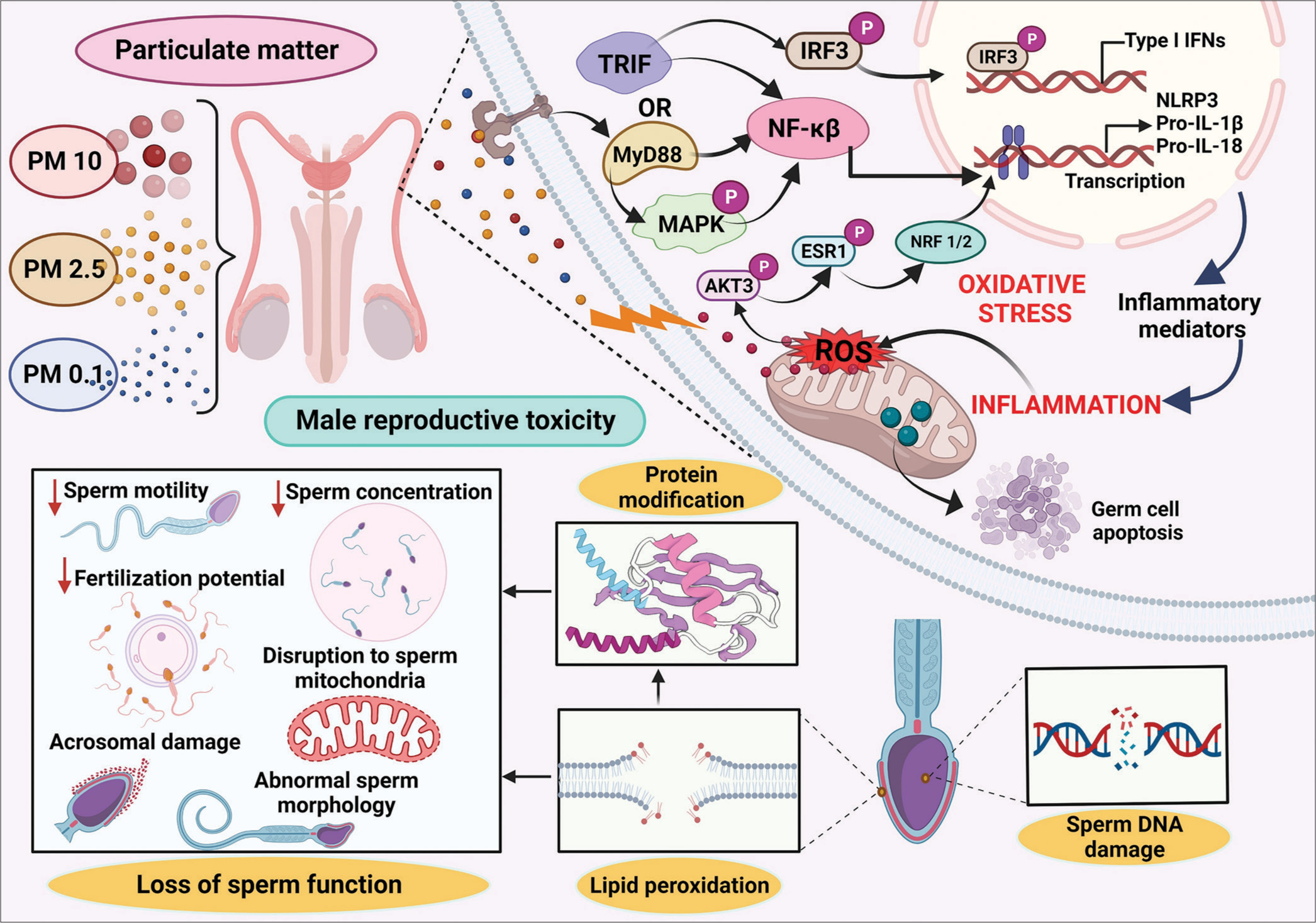
- Airborne particulate matter-associated male reproductive toxicity and involved molecular mechanisms. AKT3: Alpha serine/threonine-protein kinase 3, ESR1: Estrogen Receptor 1, IFN: Interferon, IRF: Interferon-regulatory factor, IRF3: Interferon-regulatory factor 3, MAPK: Mitogen-activated protein kinase, MyD88: Myeloid differentiation primary response 88, NF-κB: Nuclear factor-kappa B, NLRP3: Nucleotide-binding domain, leucine-rich–containing family, pyrin domain–containing-3, NRF1/2: Nuclear factor erythroid 1/2-related factor, PM: Particulate matter, Pro-IL: Pro-Interleukin, ROS: Reactive oxygen species, TRIF: Toll 1 receptor-domain-containing adaptor-inducing interferon-β
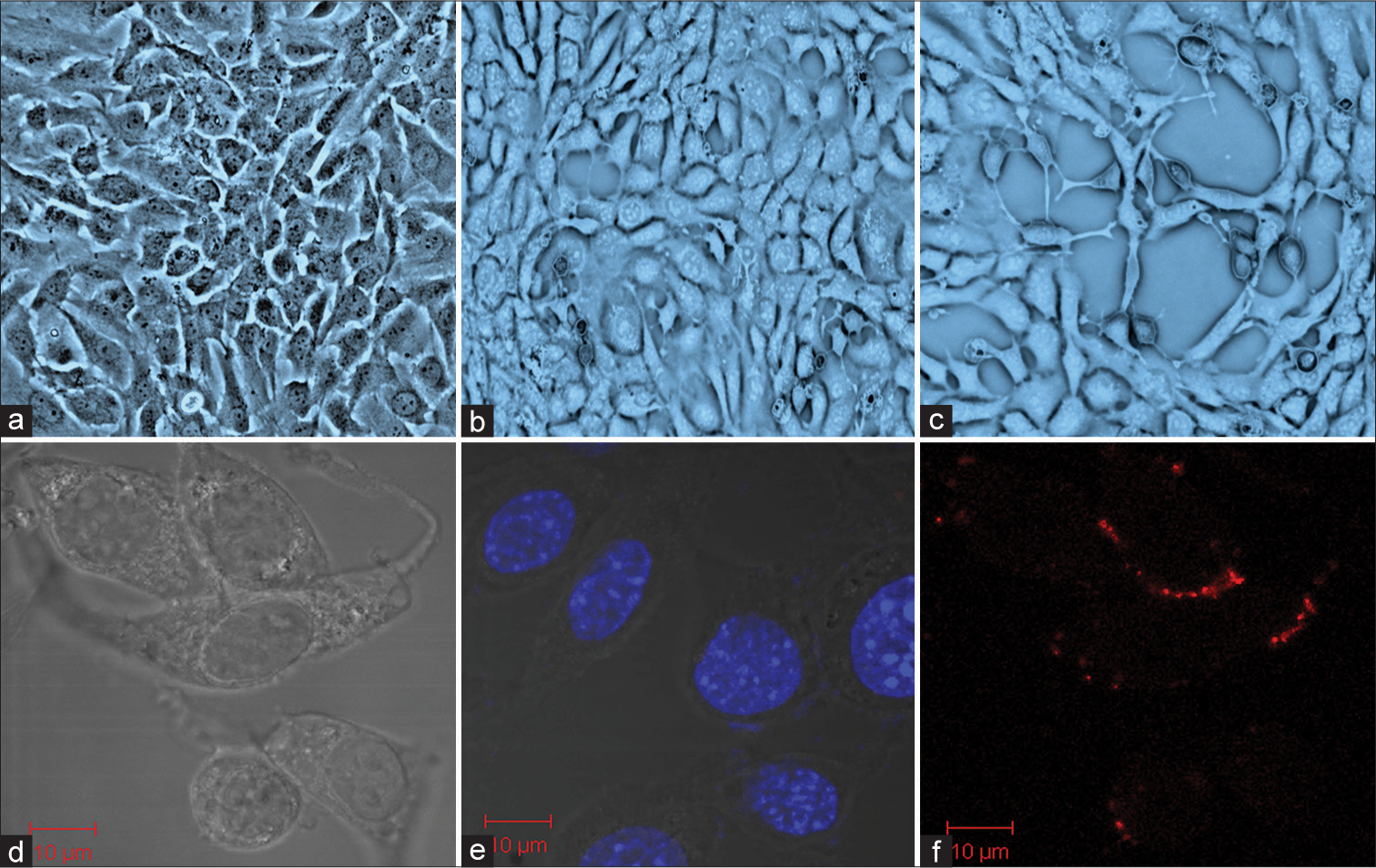
- Mouse spermatogonia epithelial (GC-1 spg) cells exposed to Ultra-fine particulate matter (UFPM) (a) control, (b) GC-1 spg cells were treated with UFPM for 24 hours, and (c) GC-1 spg cells exposed to UFPM at 72 hours. Microsopic images showing uptake of UFPM by GC1-spg cells (d) Control of GC-1 spg cells, (e) accumulation of UFPM at 24 hours, and (f) uptake of UFPM by GC-1 spg cells at 72 hours. Scale bar, 10 μm. UFPM: Ultra-fine particulate matter.
BIOMARKERS FOR MALE REPRODUCTIVE HEALTH MONITORING
A biomarker is any biological molecule capable of objective testing and assessment, indicating a physiological process. In the context of male factor infertility, biomarkers play a crucial role in evaluating a man’s capacity to father children in a precise and minimally invasive manner. Semen analysis is the most widely employed indicator for predicting male fertility potential.[27] Proteomic analyses have the potential to yield highly accurate biomarkers. Proteomics research in infertile males is still in its early stages but holds much potential. Semen analysis has already lost several novel potential biomarkers for prostate cancer.[28,29] Because it contains sperm and seminal plasma (SP), using semen for proteome research is difficult. SP comprises the prostate, seminal vesicles, and bulbourethral gland products. Fibronectin, lactoferrin, laminin, albumin, and semenogelin 1 and 2 were among the most abundant proteins.[30] Catalytic enzymatic activity was the most common molecular function of the identified proteins (60%).[29] Heparin-binding proteins represent a category of proteins proposed as potential indicators for male infertility.[31] In addition, prolactin inducible protein, found in SP, has been suggested as a biomarker for male factor infertility.[32,33] The field of metabolomics, characterized by its high-throughput technology, holds promise for rapid advancements in the discovery of real-world biomarkers and diagnostic applications. Initially, metabolomics focused on identifying changes associated with oxidative stress in the search for male fertility biomarkers. Oxidative stress induces abnormalities in spermatogenesis by increasing the synthesis and formation of ROS while impairing the antioxidant defense system. Previous studies have demonstrated that markers of oxidative stress (−CH, −NH, −OH, and −SH) impact sperm and oocyte quality, as well as embryo viability.[30] This underscores the potential of metabolomics in uncovering biomarkers that could enhance our understanding of male fertility and contribute to diagnostic advancements.
OMICS-BASED APPROACHES FOR DETECTION OF PREDICTIVE BIOMARKERS
The discovery and validation of biomarkers associated with fertility traits, encompassing genes, transcripts, proteins, and metabolites, hold considerable promise for enhancing human reproductive health.[34] Coding RNA, particularly messenger RNA (mRNA), assumes a crucial role in synthesizing functional proteins, while noncoding RNAs fulfill additional roles, with mRNA acting as a transient intermediary molecule in the information network. Proteins and metabolites are pivotal in governing the molecular pathways integral to spermatozoa and ovarian physiological processes.[35]
Genomics-based approaches
Genomics, the exploration of genomes’ structure, function, and evolution, involves using enzymes and messenger molecules to orchestrate protein production. Transcriptomics, on the other hand, delves into the comprehensive analysis of mRNA molecules within a cell, tissue, or organism, including their overall quantity and the concentration of individual RNA molecules. Proteomics takes a broader stance, encompassing the study of all proteins within a biological entity to unravel their functional and biochemical properties.[36] Genomic techniques, categorized into array-based and sequencing-based methods, are pivotal in identifying genetic connections and variants associated with complex disorders. Microarray technologies, a subset of genomic techniques, have enabled rapid assessments of spermatozoa RNAs. Utilizing these techniques, studies have revealed mRNA patterns intricately linked to critical processes such as spermatogenesis, sperm motility, anti-apoptotic mechanisms in germ cells, DNA repair, oxidative stress reduction, and histone modification. Integrating genomics, transcriptomics, and proteomics provides a holistic understanding of the intricate molecular processes governing reproductive biology, paving the way for advancements in diagnostics and therapeutic interventions.[37]
Comparative genomic hybridization (CGH)
Although the most used method for detecting copy number variations (CNVs) is CGH, it has drawbacks such as a high cost and a tight demand for DNA quality. Male infertility is mostly caused by CNV[38] [Figure 3a]. The abrupt halt in spermatogenesis, usually at the level of primary spermatocytes, is the hallmark of male infertility due to spermatogenesis maturation arrest. Array CGH uses this technique to look for deletions or duplications that could be linked to maturation arrest. We investigated a small but well-defined group of patients who had a meiotic arrest in spermatogenesis. In this patient group, it is more likely that an alteration of CNV in a single region or gene involved in spermatogenesis is the underlying cause of fertility issues.[39] The technology has been modified to use microarrays rather than metaphase chromosomes, which has made it easier to analyze more than 200,000 discrete foci. This has greatly improved the accuracy of aneuploidy analysis and increased the ability to identify small, submicroscopic microdeletions.[40] Numerous CNVs have been identified in both healthy individuals and patients with developmental delays and multiple congenital anomalies due to the development and application of high-resolution array-based comparative genome hybridization (array CGH).[41]
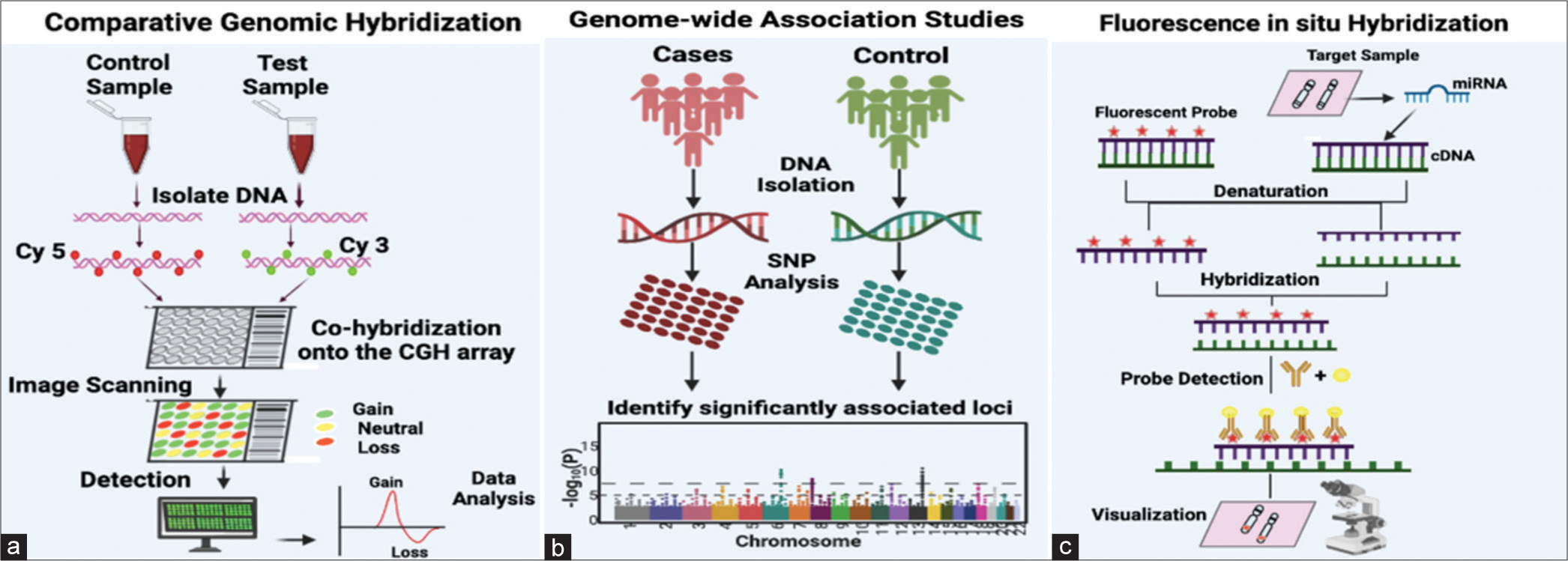
- (a) Fundamental methodology for comparative genomic hybridization analysis, wherein the control DNA sample is labeled with a red fluorescent dye, and the test DNA sample is labeled with a green, fluorescent dye. Both the labeled DNA samples, representing control and test samples, are introduced into the wells of a microarray, engaging in competitive hybridization with pre-bound complementary DNA sequences on the microarray. Subsequently, digital imaging systems measure the fluorescent signals, detecting whether there is equal/neutral hybridization, DNA dosage loss, or DNA dosage gain. The collected data are then analyzed, and plots are generated based on the outcomes of the analysis. (b) Single-nucleotide polymorphisms (SNPs) represent the genetic variants most examined in genome-wide association studies (GWAS). The diagram illustrates the steps involved in the GWAS workflow: Beginning with the collection of DNA samples from specific groups of individuals or from biobanks. Subsequent steps include genotyping using available GWAS arrays or sequencing techniques, implementing quality control measures, imputing untyped variations (SNPs) through haplotype phasing and reference populations, seeking independent replication, and finally, analyzing the outcomes with various post-GWAS analyses. (c) Fluorescence in situ hybridization, wherein a complementary nucleic acid probe is synthesized and labeled using either fluorescence or radioactivity. The labeled probe and the target DNA sequence undergo denaturation. The slide is coated with a hybridization solution containing the probe, facilitating its hybridization with the complementary target sequence. After removing any excess probe, the position of the probe can be identified through autoradiography or immunocytochemistry. Cy3: Cyanine 3; Cy5: Cyanine 5.
Genome-wide association studies (GWAS)
Numerous GWAS have investigated the involvement of single-nucleotide polymorphisms (SNPs) in idiopathic male infertility.[42] GWASs based on previous studies show that various SNPs were linked to azoospermia and oligozoospermia associated with male infertility [Figure 3b]. Another GWAS, which included 269 members of a Hutterite population in the United States, found that nine of the 41 SNPs linked to family size or birth rate were also linked to lower sperm quality and function in ethnically diverse men from Chicago. In a subsequent study conducted in 2017, a statistically significant correlation was identified between oligozoospermia and the rs12376894 proxy SNP of rs10966811. A significant association was also observed between rs10129954 and both azoospermia and oligozoospermia. The investigation revealed that rs12348 in TUSC1 was specifically linked to azoospermia and oligozoospermia, whereas rs2772579 in IZUMO3 showed no such association. Consequently, the study concluded that polymorphisms in TUSC1 and DPF3 were strongly associated with male infertility – top of the form.[43] Kyrgiafini et al., in 2020, discovered six long-non-coding RNAs (LINC02231, LINC00347, LINC02134, NCRNA00157, LINC02493, and Lnc-CASK-1) that interact with microRNAs (miRNAs) and are linked to male infertility. Moreover, they identified the targeted genes of these miRNAs and deliberated on their functions in spermatogenesis and fertilization.[44,45]
Fluorescence in situ hybridization (FISH)
The FISH is becoming more and more popular in the field of genomics to identify specific chromosomal regions that may be linked to poor semen quality and fertility, even though cytogenetic karyotyping of peripheral blood lymphocytes can provide basic information about chromosome number and structure.[46,47] FISH is a very accurate and dependable method that uses fluorochrome-labeled chromosome-specific probes to analyze a large quantity of sperm [Figure 3c]. Using the multi-probe FISH technique for sperm analysis, the numerical analysis of chromosomes has been added as a standard test for the screening of infertile couples.[48] A retrospective study was carried out to investigate sperm defects using FISH, which compared samples from sterile patients and healthy people. Researchers concluded that oligozoospermia, asthenozoospermia, and teratozoospermia patients had abnormal FISH results.[49] In spermatogenesis, FISH gives a rough idea of the frequency of chromosomal abnormalities.[50] FISH technology has made it easier to study sperm from patients who have severe spermatogenesis defects. The analysis of sperm aneuploidy can now be accelerated using automated FISH imaging and analysis technology.[40]
Transcriptomics-based approaches
Transcriptomics technologies encompass methods designed to explore an organism’s transcriptome, which constitutes the total RNA transcripts within that organism. An organism’s genetic information is encoded in its genome’s DNA and expressed through transcription. While mRNA is a temporary intermediary molecule in the information network, noncoding RNAs fulfill additional functions. A transcriptome captures the entirety of transcripts present in a cell, offering a snapshot at a specific moment in time. Initial efforts to analyze the complete transcriptome emerged in the early 1990s, and since then, technical advancements have propelled transcriptomics into a widely recognized discipline.[51] The field of transcriptomics has evolved through a series of technological breakthroughs that have significantly transformed the discipline. Despite their presence in sperm and transmission to the egg, the biological significance of RNAs generated during the pre-fertilization phase of transcription has been a subject of inquiry. Emerging data suggest that miRNAs (miRs) and Piwi-interacting (piRNAs) may play pivotal roles in early embryonic development and serve as indicators of male fertility.
RNA sequencing (RNA-Seq)
The presence and quantity of RNA in sperm have been investigated using RNA-Seq and other methodologies in various species, such as bovine, swine, and avian sperm. Notable impacts on fertility have been observed, including variations in mRNA transcripts, miRNA, and piRNA[52] [Figure 4a]. The previous research identified 805 transcripts unique to the higher fertility group, 2944 transcripts exclusive to the lower fertility population, and 2422 transcripts shared between the two populations. The higher-fertility bulls had larger regulatory transcripts for growth and protein kinase activity, according to an additional study based on GO keywords. As it turns out, the gene for the electron transport chain’s last stage, leading to ATP production, was specifically associated with lower sire fertility, and that gene, COX7C, was shown to be the culprit, as higher levels of COX7C seen in the lower fertility sires were due to improper translation of this transcript and, as a result, impaired mitochondrial activity in the latter stages of spermatogenesis[53] It is common to find that the quality and fertility of boar semen are lower in the summer and higher in the winter months, respectively. This study utilized deep sequencing to identify 34 genes and seven miRNAs with a distinct distribution linked to oxidative stress, DNA damage, and autophagy in the boar ejaculate transcriptome. A comparison of gene expression microarray analyses revealed 21,639 transcripts in chicken testis and 17,542 transcripts in sperm RNA transcriptomes. The sperm and testis shared 66.54% of the transcripts. However, the sperm alone accounted for 33.45% of the transcripts.[54] The retention of RNA in spermatozoa is comparable to that of mRNA during oogenesis, where substantial amounts of mRNA are retained until transcription is completed. Mature oocytes depend on the translational reservoirs of maternal mRNA for protein synthesis. Both gametes undergo the same translation modification, with synchronized alterations in the poly-A-tail. This synchronization occurs through active interactions between appropriate RNA-binding proteins and untranslated regions of RNA. Within the spermatozoa ejaculate, two distinct populations of mRNA exist, each serving its specific function: a centriolar population that contributes to embryonic maintenance and developmental processes and a nuclear population that assists in chromatin repackaging or the transitional replacement of “spent” proteins.
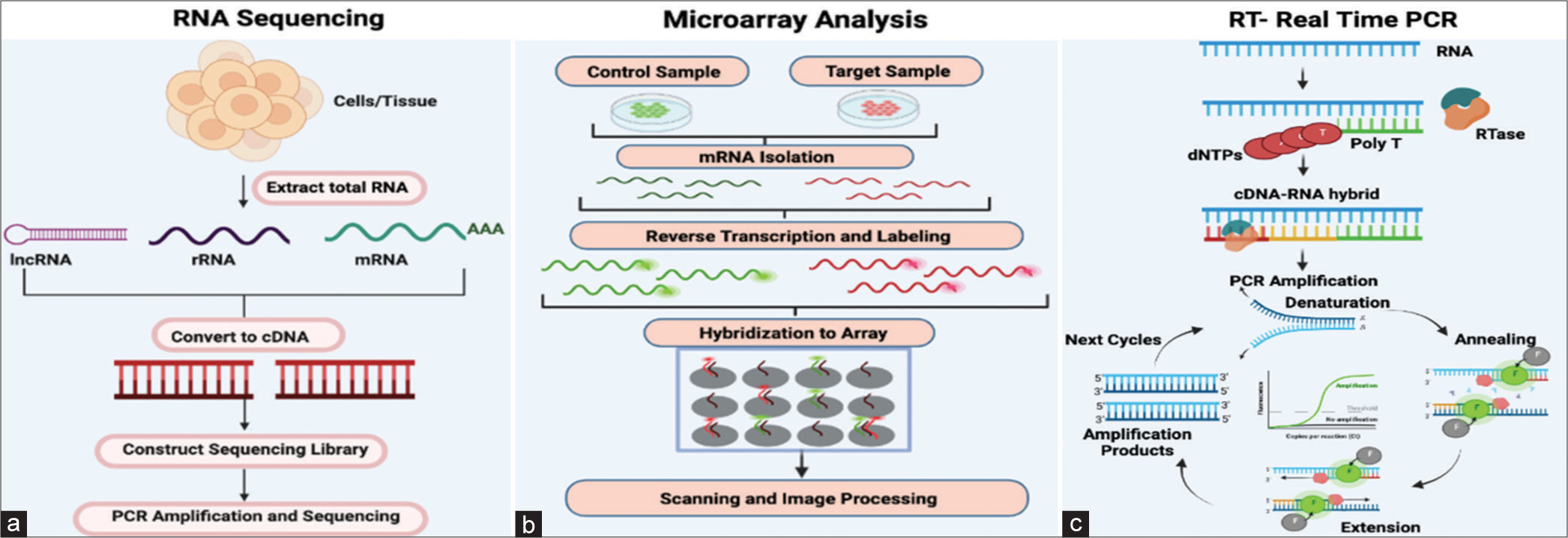
- (a) This figure illustrates the process of RNA sequencing, detailing the key steps. Initially, cells or tissues are utilized to extract total RNA. Subsequently, specific protocols are employed to isolate subsets of RNA molecules. Examples of these protocols are ribo-depletion to eliminate ribosomal RNAs and poly-A selection to enrich for polyadenylated transcripts. Then, reverse transcription transforms the RNA into cDNA and sequencing adaptors are then ligated to the ends of the cDNA segments. The RNA sequencing library is ready for sequencing after polymerase chain reaction (PCR) amplification. (b) Microarray analysis is carried out by collecting both control and target samples from which mRNAs are isolated. Both mRNA control and test samples are then converted into cDNAs which are then labelled with fluorescent probes. In the figure, control cDNA sample is labeled with green, fluorescent dye and test cDNA sample with red fluorescent dye. The two samples are then mixed and allowed to bind to the microarray slide (hybridization). Subsequently, the microarray is subjected to scanning following hybridization to quantify the expression of each gene imprinted on the slide. The gene expression profiles are then derived from the analysis of the collected data. (c) This diagram represents the fundamental stages of real-time PCR. Initially, RNA is isolated, and both its quantity and integrity are evaluated. Then, cDNA is synthesized and subsequently utilized as a PCR template in a two-step process. The following stage amplifies and detects genes using fluorescent reporters and a PCR reaction. A normalization factor must be computed for every sample to normalize findings with numerous housekeeping genes. dNTPs: Dinucleotide triphosphates; RTase: Reverse transcriptase.
Microarray analysis
Two modern techniques used in the field are microarrays and RNA-Seq. Microarrays measure a specific set of present sequences, while RNA-Seq captures all sequences using high-throughput sequencing [Figure 4b]. A study was conducted to identify Y-chromosome microdeletions using molecular cytogenetics. The study used CGH chromosome microarray analysis (CMA) and found that 52% of cases had additional cytogenetic anomalies that were clinically significant. These anomalies included six cases of 46-year-old males with XX genotypes, one case of an isodicentric Y, two cases of a dicentric Y, and three cases of terminal Yq deletions.[55]
Real-time polymerase chain reaction (PCR)
When evaluating male infertility caused by a low sperm count or no sperm, doctors usually conduct two tests: a karyotype and a Y-chromosome microdeletion analysis. These tests help determine the cause of infertility. The latter is commonly performed using a multiplex PCR assay. Using multiplex PCR, it is possible to detect approximately 20 Y-chromosome loci, including the azoospermia factor (AZF) regions. This is achieved by amplifying sequence-tagged sites along the chromosome[56] [Figure 4c].
Proteomics-based approaches
The significant content of tissue-specific proteins in SP makes it a potential source of biomarkers for assessing male fertility. Proteomic and biomarker discovery technologies have advanced over the past few decades, making it possible to identify thousands of high, medium, and low-abundance proteins in large quantities using various methods, including mass spectrometry (MS), liquid chromatography, and basic electrophoresis. In the initial research on groundbreaking protein biomarkers conducted in the 1940s, just four unique protein separations were found. Identification of about 2300 seminal proteins was performed in both fertile and infertile men using MS techniques.[57] By examining these cohorts, researchers hope to create natural fertility biomarker panels for the diagnosis and treatment of male infertility. SP protein levels and the outcomes of routine semen analysis have been compared in several studies. Reaching asthenozoospermic (AS) men to normal men, there was a decrease in the expression of some proteins and an increase in the expression of others. The expression of DJ-1-a, a regulator of oxidative stress, was found to be lower in males with AS. This suggests that the regulation of oxidative stress may be compromised in men with decreased sperm motility. In a study of 202 ejaculates, A study of 202 ejaculates revealed a positive correlation between seminal prostaglandin D synthase (PTGDS) concentration and sperm count, motility, and normal morphology. The levels of PTGDS decreased in males with oligozoospermia, azoospermia, and those who underwent vasectomy, indicating a connection between reduced PTGDS and testicular occlusion.[57] Rolland et al. conducted a similar study and identified 699 proteins in the seminal proteomes of males with normal, azoospermic, and post-vasectomy sperm.[58] They showed that TKTL1, LDHC, and PGG2 were easily identifiable in normal SA males but absent or barely detectable in the post-vasectomy and azoospermia groups. In a second cohort comparison, eight different proteins were found to be upregulated in azoospermic men: fibronectin, prostatic acid phosphatase, prolactin-inducible protein, beta-2 microglobulin, proteasomal subunit-alpha type-3, galectin-3-binding protein, and cytosolic non-specific dipeptidase. Proteome research has improved our understanding of male infertility, but diagnostic tools beyond semen analysis are still lacking.[59] Proteomics is the comprehensive analysis of proteins found in a tissue or cell. In proteomics, shotgun or bottom-up techniques are the most common, and they may detect more than 1000 proteins in a single sample. In the study of male infertility, semen is the biological fluid that is most frequently utilized to assess men’s reproductive health. The cellular and non-cellular components of spermatozoa and the SP present in sperm can be analyzed using proteomics techniques. Proteomics can be used to explain functional protein pathways in various illnesses, identify several diagnostic signs, and better understand pathogenic mechanisms. Operations on omics have proven easy to perform since controls and unaffected patients are recruited. However, they have several drawbacks because they are predicated on an implausible connection between a putative gene, mRNA, or protein and disease, which is constrained by the disorder’s understanding.[60] Since a protein regulates each of these physiological processes, variations in the genes that encode these proteins or their translation could be potential fertility predictors. Metabolites, epigenetic modifications, and small noncoding RNAs might all be valuable fertility indicators in the future. Abundant molecular analysis techniques and technology are now accessible to investigate the genome, transcriptome, proteome, metabolome, and epigenetic factors linked to male fertility. By exploiting these variations, they can influence heredity, embryo development, fertility, and infertility. Using a data mining technique, Panner et al. discovered 6198 proteins in human spermatozoa, compared to 2064 proteins in SP. Spermatozoa rely on proteins to perform their roles because they are transcriptionally and translationally inert. [61] Modifications in the SP proteome and the spermatozoa cause defects in the normal physiological processes of spermatozoa. These abnormalities have been related to high oxidative stress, varicocele, idiopathic infertility, and unexplained male infertility (UMI), among other male infertility illnesses. Hyperactivation, capacitation, acrosome reaction, and fertilization are biomarkers for male infertility. Proteomics research could aid in developing new techniques for identifying novel biomarkers for a more accurate clinical diagnosis and treatment of male infertility. The discovery of new pathways and molecular mechanisms in male infertility could aid in developing more unique therapeutic techniques in this area. In the examination of male fertility/infertility, SP is a good sample for proteome analysis. The epididymis and auxiliary sexual glands contribute to SP, an acellular fluid conglomerate. Many proteins in human SP are required for the proper fertilization of the egg by spermatozoa. Male infertility disorders such as varicocele, idiopathic infertility, UMI, excessive oxidative stress, and testicular cancer are associated with anomalies in spermatozoa’s normal physiological activities, which are brought about by changes in the spermatozoa and SP proteome. Key proteins associated with sperm activities, such as hyperactivation, capacitation, acrosome response, and fertilization process, can be used as non-invasive indicators in the pathophysiology of male infertility.[61]
Two-dimensional gel electrophoresis (2DE)
In the study of proteomics, 2DE is an essential tool. Often referred to as the proteomic “workhorse,” this ancient approach uses isoelectric focusing in the first dimension, and SDS PAGE in the second dimension is used to enable the separation of complicated protein mixtures [Figure 5a]. Stains that are fluorescent or visible can be used to visualize protein profiles. Finding biomarkers in different illness states is typically its use. Using 2DE, SP from infertile patients was compared to healthy samples. Using two-dimensional electrophoresis, the pathological conditions in the patients were identified.[62] When used in quantitative proteomics, two-dimensional difference gel electrophoresis (2D-DIGE) allows for analyzing different research groups within the same experimental design.[63]
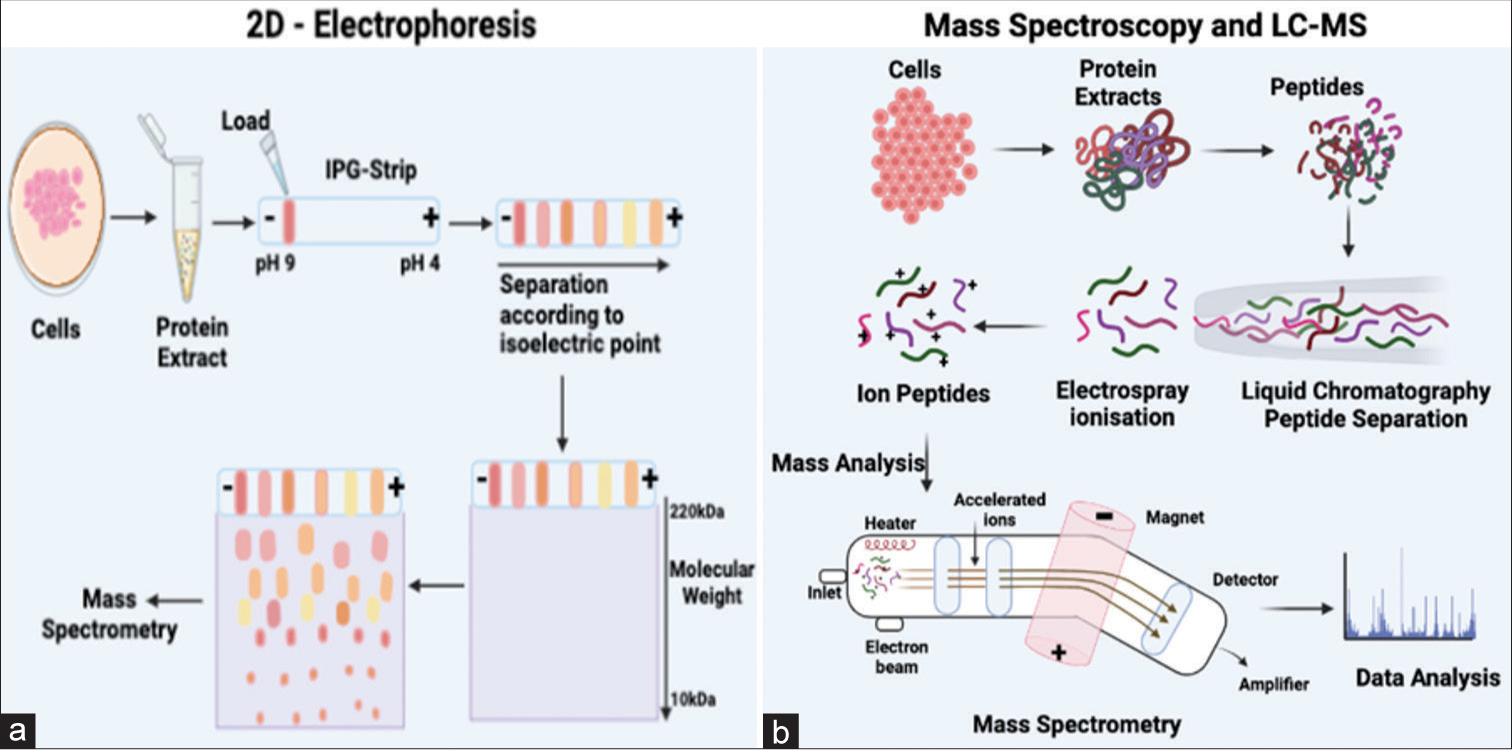
- (a) The 2D electrophoresis process employs two distinct steps to segregate proteins. Initially, the proteins undergo separation based on their isoelectric point through isoelectric focusing, followed by SDS-PAGE, which separates proteins based on their molecular weights. The outcomes obtained from this process are subsequently analyzed using mass spectrometry. (b) The figure shows the basic steps of mass spectroscopy and liquid chromatography (LC)-mass spectrometry methodologies. The interaction of the compounds with the stationary and mobile phases determines how the sample components are separated in LC, and the degree of compound separation is influenced by the affinity of each chemical for the mobile phase. Separated compounds are then delivered into the mass spectrometer for mass analysis after they elute from the column, dissolved into the gas phase, and ionize at an ionization source after chromatographic separations. IPG: Immobilized pH gradient; LC: Liquid chromatography; SDS-PAGE: Sodium dodecyl-sulfate polyacrylamide gel electrophoresis.
Mass spectroscopy (MS)
MS works on all kinds of molecules, including proteins and lipids. Separating peptides based on their mass-to-charge ratio is essential to the proteomics MS principle. An ionizer that blasts them with hydrogen ions results in charged proteins traveling through the mass analyzer and detector, which is connected to a computer program.[64] Gel-based proteomics investigations involve extracting the protein-containing region from the gel, digesting the sample with trypsin, which cleaves proteins on the C-terminal side of arginine or lysine, and eluting the proteins from acrylamide[65] [Figure 5b]. These peptides are then subjected to matrix-assisted laser desorption/ionization-time-of-flight (TOF), producing a series of peaks, each describing the molecular mass of a single peptide in the mixture. Proteomic application of MS is constantly growing as more improved instruments and smarter ways to couple them become available.[66] The most commonly used mass analyzers for protein biochemistry applications are TOF, triple-quadrupole, quadrupole-TOF, ion-trap, and hybrid or bitmap instruments. Complex protein mixtures can be studied using MS-based approaches. The mass-to-charge ratio (m/z) of ions formed from peptides and proteins within the sample, which produces precise mass values, is measured using this method. These methods are the most often used to ascertain the protein concentration of seminal fluid.
Liquid chromatography-mass spectrometry (LC-MS)
The hyphenated technology is known as LC-MS integrates the separation capabilities of LC with the detection precision of MS. This analytical chemistry technique, also referred to as LC-MS or high-performance liquid chromatography (HPLC)-MS, combines the physical separation capabilities of liquid chromatography (or HPLC) with the mass analysis capabilities of MS. The LC-MS system comprises a chromatographic column, an ionization source, a mass analyzer, and a detector. LC-MS is a versatile technology known for its excellent sensitivity and selectivity, rendering it suitable for various applications. In drug discovery and proteomics research, LC/MS has become a pivotal tool. It facilitates activities such as target protein characterization and biomarker development.
Metabolomics-based approaches
Metabolomics is characterized as investigating cells, tissues, or biological fluids by scrutinizing as many metabolites as possible. Metabolites, which represent the ultimate outcomes of cellular processes, can be conceptualized as conclusive responses to physical and environmental alterations.[67,68] A biological organization’s “metabolome” comprises the sequences of metabolites it produces. Metabolomics is a strong systems biology tool for studying global and dynamic changes in metabolism. By testing body fluids (e.g., plasma) and tissue, metabolic profiling can detect various endogenous metabolites, including organic acids, amino acids, fatty acids, sugar, cholesterol, and other compounds (e.g., testicular tissue). Optical and non-optical spectroscopic techniques, including near infrared [Figure 6a], Raman spectroscopy, LC-MS, gas chromatography-mass spectrometry (GC-MS), and nuclear magnetic resonance (NMR) spectroscopy, are commonly employed in metabolomics investigations. The choice of metabolomics study directly influences the selection of instrumentation. Infertile males experience changes in their sperm and seminal fluid proteins that are similar to those associated with aging. These changes include mitochondrial dysfunction, DNA instability, oxidative stress, protein misfolding, and intracellular mistrafficking. In addition, there are changes in processes specifically related to reproduction, such as sperm-egg recognition/acrosome reaction, embryo and morula development, blastocyst implantation, and DNA methylation involved in erectile dysfunction.
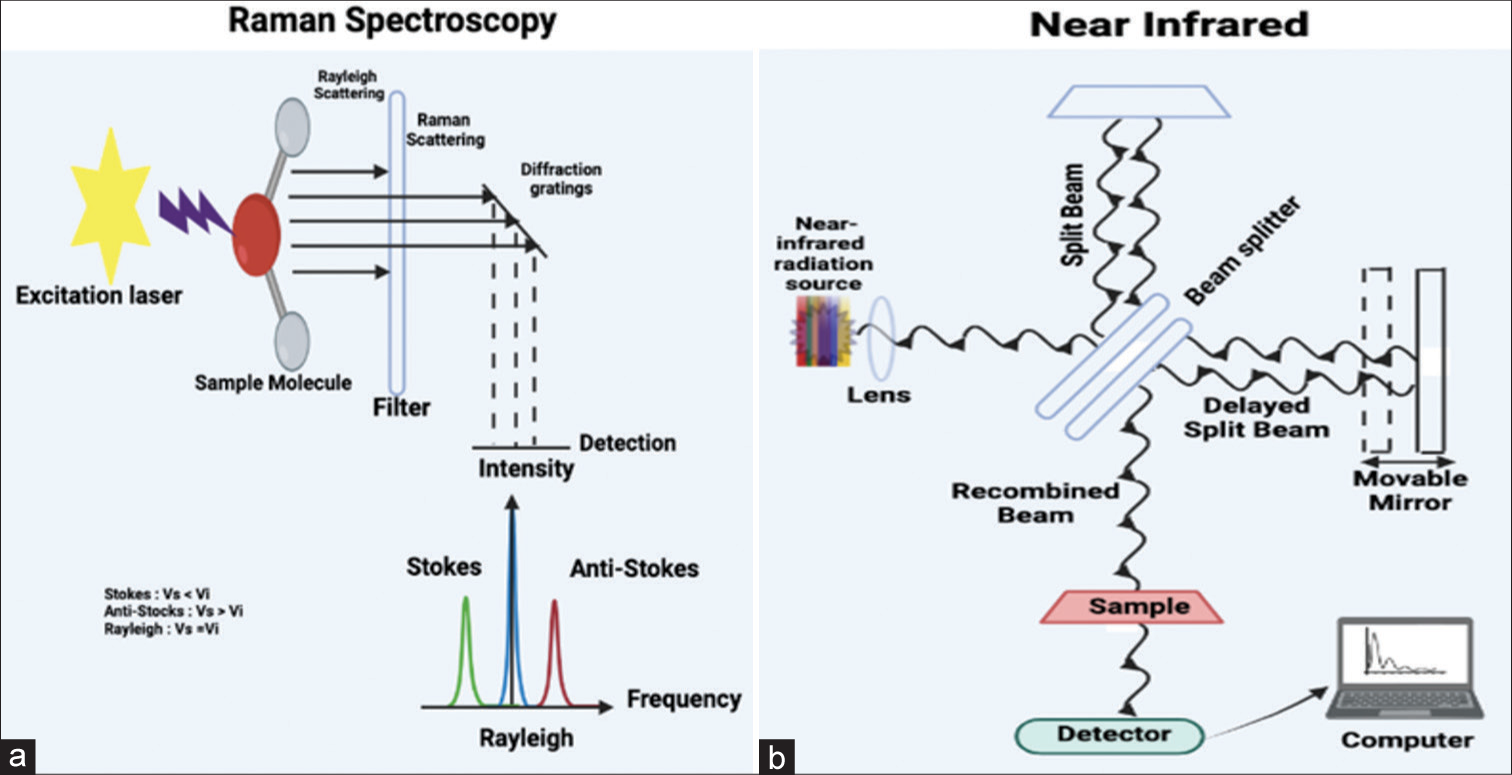
- (a) A Raman spectrum is made up of several peaks that represent the wavelength position and strength of the dispersed Raman light. Every peak is associated with a distinct molecular bond vibration. These can be individual bonds such as C–C, C=C, N–O, or C–H, or groups of bonds such as the breathing mode of the benzene ring, polymer chain vibrations, and lattice modes. (b) The diagram delineates the functioning of near-infrared spectroscopy, a technique that utilizes the near-infrared portion of the electromagnetic spectrum ranging from approximately 700–2500 nanometers. In this spectroscopy, near-infrared reflectance spectra are employed to rapidly determine the characteristics of a material without inducing any alterations to the sample. This is achieved by measuring the amount of light scattered off and transmitted through the material.
NMR spectroscopy
New research utilizing (1)H NMR-based metabolomic analysis in males with various infertility criteria, such as oligozoospermia, azoospermia, and teratozoospermia, has unveiled significant differences between fertile and infertile groups. Employing the same technique, the metabolome of males exhibiting reduced sperm motility was also explored, revealing alterations in metabolic pathways involving fat, cholesterol, nucleosides, and phospholipids. The latest urinary metabolome study on oligozoospermia conducted by Zhang and colleagues identified reduced levels of aspartic acid, leucylproline, and acylcarnitines, alongside elevated levels of methylxanthine and adenine, strongly associated with the risk of decreased sperm count (oligozoospermia). Increased concentrations of lipids related to cytidine triphosphate (CTP), uridine triphosphate, guanosine triphosphate (GTP) production, arachidonic acid metabolism, and sterol biosynthesis were found, implying that signal transduction in these males is likely altered. Recent Raman spectroscopy investigations for metabolomics fingerprinting have exposed an imbalanced generation of ROS linked to oxidative stress in men with idiopathic infertility [Figure 6b]. In a study by Zhang and colleagues utilizing (1)H NMR spectroscopy, an elevation in oxysterols such as 5-cholesterol and 7-ketocholesterol was identified in the SP of AS patients, underscoring the significance of processes induced by oxidative stress in the degradation of semen quality.[69,70]
Gas chromatography
GC-MS was employed for untargeted metabolomics profiling, enabling the differentiation of a unique metabolomic profile. Using 2D-DIGE in quantitative proteomics facilitates analyzing various study groups within the same experimental design.[71] Metabolomics, as a field of study, is characterized by quantitatively depicting all endogenous molecular metabolites in tissues and cells, employing diverse spectroscopy and analytical techniques. Metabolic profiling is to identify the metabolic products linked to pathological and physiological conditions.[72] Human sperm cells from individuals with normozoospermic and idiopathic asthenozoospermia were examined using GC-MS to determine their metabolomes.[73]
CONCLUSION
The exploration of infertility’s global impact emphasizes the critical need to address its profound challenges affecting families and communities worldwide. Variations in reproductive lifespan and fertility present profound implications for health, population dynamics, and aging. The escalating evidence linking environmental air pollution, particularly PM, to reproductive health underscores the urgency of understanding and mitigating its detrimental effects on male reproductive health, offering insights into the molecular pathways underlying male reproductive health, including NF-kb, MAPK, and phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathways that alter the blood-testes barrier. Various omics techniques, such as CGH, fluorescent in situ hybridization, GWAS, RNA-Seq, microarray analysis, reverse transcriptase real-time PCR, LC-MS, 2D-gel electrophoresis, RAMAN spectroscopy, near infrared, and NMR, play crucial roles in understanding and evaluating reproductive health biomarkers. This comprehensive review highlights the current advancements in omics as an opportunity to study pathological mechanisms and investigate molecular pathways crucial for reproductive health. The elucidation of biomarkers and molecular pathways holds promise for enhancing diagnostics and therapeutic interventions to address the global challenge of male infertility linked to environmental factors like air pollution. Through the process of epigenetics, diverse environmental pollutants may alter the reproductive health status of an individual and their progeny. Toxins in the environment could affect human reproductive systems. PM, specifically PM0.1, emerges as a significant factor, inducing inflammation, oxidative stress, DNA variations, epigenetic changes, and endocrine disruption factors intricately linked to male reproductive health. The identification and validation of biomarkers associated with PM exposure, spanning genes, transcripts, proteins, and metabolites, offer immense potential for diminishing reproductive efficiency. Genetic factors contribute to infertility, prompting extensive research to uncover implicated genes. State of the art omics techniques, including genomics, transcriptomics, proteomics, and metabolomics, provide a novel opportunity to delve into the molecular pathways governing male reproductive health.
FUTURE PERSPECTIVES
Omics-based technologies have emerged as promising methodologies for determining the molecular bases of male infertility. The utilization of high-throughput systems for protein and metabolite detection, coupled with advanced bioinformatics software for their analysis, facilitates the identification of biomarkers for male infertility diagnosis, prognosis, and prediction of reproductive outcomes.[74,75] The understanding of the molecular alterations associated with male infertility can provide a basis for the development of effective treatments with significant implications for the therapeutic management of infertile patients. The advancement of cutting-edge technologies is revolutionizing our comprehension of the effects of airborne PM on male reproductive health. Advanced imaging techniques, such as high-resolution microscopy and in vivo imaging, offer real-time visualization of PM interactions within the male reproductive system. Single-cell sequencing, a breakthrough in genomics, allows for the precise identification of PM-induced genetic alterations in male germ cells. In the realm of proteomics, MS and advanced chromatographic methods provide detailed insights into PM-induced protein changes, offering a better understanding of molecular pathways affecting male fertility. Metabolomics, utilizing technologies such as NMR and MS, explores the metabolic signatures associated with PM exposure, linking environmental pollutants to disruptions in crucial cellular processes.
Furthermore, biomarker discovery, facilitated by high-throughput omics technologies, identifies specific genes, transcripts, proteins, and metabolites indicative of PM-related reproductive damage. In addition, wearable technologies and environmental sensors enable real-time monitoring of personal exposure to air pollutants. The integration of this data with health outcomes allows for a comprehensive understanding of the cumulative impact of air pollution on male reproductive health. Together with systems biology approaches, leveraging computational models and artificial intelligence may help to integrate multi-omics data to identify key molecular pathways and predict adverse reproductive outcomes. Advanced data analytics and artificial intelligence might enhance our ability to decipher complex omics datasets, uncovering subtle patterns and associations crucial for understanding the nuanced impact of airborne PM on male reproductive health. These interdisciplinary technologies collectively will propel research forward, shedding light on the intricate mechanisms underlying PM-mediated reproductive toxicity and opening avenues for targeted interventions and preventive strategies. The precision and depth of information garnered from these technologies hold the key to early detection of subtle molecular changes, allowing for proactive interventions and personalized medical strategies. Integration of multi-omics data, coupled with advanced analytics and machine learning, promises a deeper understanding of the intricate molecular mechanisms underpinning the effects of air pollution on reproductive health. This wealth of information is not only instrumental in refining regulatory standards and environmental policies but also sets the stage for a new era of personalized reproductive medicine, where interventions are tailored based on an individual’s unique molecular profile in the context of environmental exposures. Ultimately, the omics revolution stands to usher in a future where we can safeguard reproductive health with unprecedented precision and insights.
Ethical approval
The Institutional Review Board approval is not required.
Declaration of patient consent
Patient’s consent was not required as there are no patients in this study.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript, and no images were manipulated using AI.
Financial support and sponsorship
RK and PR express their gratitude toward the Indian Council of Medical Research (ICMR), Department of Health Research (DHR), Ministry of Health and Family Welfare, Government of India, New Delhi, for providing financial support in the form of Senior Research Fellowship (SRF).
References
- Reproductomics: An impending driver for exposome research. J Reprod Health Med. 2022;3:1.
- [CrossRef] [Google Scholar]
- Environmental impact on reproductive health: Can biomarkers offer any help? J Reprod Infertil. 2017;18:336-40.
- [Google Scholar]
- Does air pollution play a role in infertility? A systematic review. Environ Health. 2017;16:82.
- [CrossRef] [PubMed] [Google Scholar]
- Air pollution-induced epigenetic changes: Disease development and a possible link with hypersensitivity pneumonitis. Environ Sci Pollut Res Int. 2021;28:55981-6002.
- [CrossRef] [PubMed] [Google Scholar]
- Particulate matter air pollution: Effects on the cardiovascular system. Front Endocrinol (Lausanne). 2018;9:680.
- [CrossRef] [PubMed] [Google Scholar]
- Air pollution associated epigenetic modifications: Transgenerational inheritance and underlying molecular mechanisms. Sci Total Environ. 2019;656:760-77.
- [CrossRef] [PubMed] [Google Scholar]
- Air pollution exposure and bladder, kidney and urinary tract cancer risk: A systematic review. Environ Pollut. 2020;267:115328.
- [CrossRef] [PubMed] [Google Scholar]
- A review on the human health impact of airborne particulate matter. Environ Int. 2015;74:136-43.
- [CrossRef] [PubMed] [Google Scholar]
- Elemental composition of PM2.5 and PM10 and health risks assessment in the industrial districts of Chelyabinsk, South Ural Region, Russia. Int J Environ Res Public Health. 2021;18:12354.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative profiling of epigenetic modifications among individuals living in different high and low air pollution zones: A pilot study from India. Environ Adv. 2021;4:100052.
- [CrossRef] [Google Scholar]
- Mapping the mitochondrial regulation of epigenetic modifications in association with carcinogenic and noncarcinogenic polycyclic aromatic hydrocarbon exposure. Int J Toxicol. 2020;39:465-76.
- [CrossRef] [PubMed] [Google Scholar]
- Emerging role of mitochondria in airborne particulate matter-induced immunotoxicity. Environ Pollut. 2021;270:116242.
- [CrossRef] [PubMed] [Google Scholar]
- Impact of air pollution on asthma outcomes. Int J Environ Res Public Health. 2020;17:6212.
- [CrossRef] [PubMed] [Google Scholar]
- Integrated mitoepigenetic signalling mechanisms associated with airborne particulate matter exposure: A cross-sectional pilot study. Atmos Pollut Res. 2022;13:101399.
- [CrossRef] [Google Scholar]
- Exposure to ultrafine particulate matter induces NF-κβ mediated epigenetic modifications. Environ Pollut. 2019;252:39-50.
- [CrossRef] [PubMed] [Google Scholar]
- Air pollution and female fertility: A systematic review of literature. Reprod Biol Endocrinol. 2018;16:117.
- [CrossRef] [PubMed] [Google Scholar]
- Prenatal exposure to environmental prooxidants induces mitochondria-mediated epigenetic changes: A cross-sectional pilot study. Environ Sci Pollut Res Int. 2022;29:74133-49.
- [CrossRef] [PubMed] [Google Scholar]
- Human health effects of air pollution. Environ Pollut. 2008;151:362-7.
- [CrossRef] [PubMed] [Google Scholar]
- Ultrafine particulate matter impairs mitochondrial redox homeostasis and activates phosphatidylinositol 3-kinase mediated DNA damage responses in lymphocytes. Environ Pollut. 2018;234:406-19.
- [CrossRef] [PubMed] [Google Scholar]
- Biological effects and dose-response assessment of diesel exhaust particles on in vitro early embryo development in mice. Toxicol Sci. 2010;117:200-8.
- [CrossRef] [PubMed] [Google Scholar]
- Environmental contaminants and human infertility: Hypothesis or cause for concern? J Toxicol Environ Health B Crit Rev. 2008;11:162-76.
- [CrossRef] [PubMed] [Google Scholar]
- Impact of air pollution on fertility: A systematic review. Gynecol Endocrinol. 2015;31:7-13.
- [CrossRef] [PubMed] [Google Scholar]
- Mitochondrial anomalies: Driver to age associated degenerative human ailments. Front Biosci (Landmark Ed). 2016;21:769-93.
- [CrossRef] [PubMed] [Google Scholar]
- Fetal nucleic acids in maternal plasma: From biology to clinical translation. Front Biosci (Landmark Ed). 2018;23:397-431.
- [CrossRef] [PubMed] [Google Scholar]
- Applying the exposome concept in birth cohort research: A review of statistical approaches. Eur J Epidemiol. 2020;35:193-204.
- [CrossRef] [PubMed] [Google Scholar]
- Oxidation-reduction potential of semen: What is its role in the treatment of male infertility? Ther Adv Urol. 2016;8:302-18.
- [CrossRef] [PubMed] [Google Scholar]
- Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7:R40.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization of isocyanate-induced male germ-line genomic instability. J Environ Pathol Toxicol Oncol. 2010;29:213-34.
- [CrossRef] [PubMed] [Google Scholar]
- Proteomic analysis of heparin-binding proteins from human seminal plasma: A step towards identification of molecular markers of male fertility. J Biosci. 2009;34:899-908.
- [CrossRef] [PubMed] [Google Scholar]
- Proteomic analysis of seminal plasma in men with different spermatogenic impairment. Andrologia. 2012;44:256-64.
- [CrossRef] [PubMed] [Google Scholar]
- Human serum albumin as a new interacting partner of prolactin inducible protein in human seminal plasma. Int J Biol Macromol. 2012;50:317-22.
- [CrossRef] [PubMed] [Google Scholar]
- A pragmatic and translational approach of human biomonitoring to methyl isocyanate exposure in Bhopal. Indian J Med Res. 2012;135:479-84.
- [Google Scholar]
- mRNA-based therapeutics: Powerful and versatile tools to combat diseases. Signal Transduct Target Ther. 2022;7:166.
- [CrossRef] [PubMed] [Google Scholar]
- Genome survey of chromatin-modifying enzymes in threespine stickleback: A crucial epigenetic toolkit for adaptation? Front Mar Sci. 2019;6:721.
- [CrossRef] [Google Scholar]
- Nanophotonic biosensors as point-of-care tools for preventive health interventions. Nanomedicine (Lond). 2020;15:1541-4.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of comparative genomic hybridization technologies across microarray platforms. J Biomol Tech. 2009;20:135-51.
- [Google Scholar]
- Array comparative genomic hybridization in male infertility. Hum Reprod. 2012;27:921-9.
- [CrossRef] [PubMed] [Google Scholar]
- The clinical implementation of sperm chromosome aneuploidy testing: Pitfalls and promises. J Androl. 2008;29:124-33.
- [CrossRef] [PubMed] [Google Scholar]
- Interpretation of array comparative genome hybridization data: A major challenge. Cytogenet Genome Res. 2011;135:222-7.
- [CrossRef] [PubMed] [Google Scholar]
- Single nucleotide polymorphisms and idiopathic male infertility in GWAS: A meta-analysis. Eur J Public Health. 2021;31(Suppl 3):164-855.
- [CrossRef] [Google Scholar]
- Association study between single-nucleotide variants rs12097821, rs2477686, and rs10842262 and idiopathic male infertility risk in Serbian population with meta-analysis. J Assist Reprod Genet. 2020;37:2839-52.
- [CrossRef] [PubMed] [Google Scholar]
- The role of long noncoding RNAs on male infertility: A systematic review and in silico analysis. Biology (Basel). 2022;11:1510.
- [CrossRef] [PubMed] [Google Scholar]
- Genome-wide association studies on endometriosis and endometriosis-related infertility. BioRxiv 2018:401448.
- [CrossRef] [Google Scholar]
- Cytogenic and molecular studies of male infertility in cases of Y chromosome balanced reciprocal translocation. Mol Med Rep. 2017;16:2051-4.
- [CrossRef] [PubMed] [Google Scholar]
- Aneuploidy in human spermatozoa: FISH analysis in men with constitutional chromosomal abnormalities, and in infertile men. Reproduction. 2001;121:655-66.
- [CrossRef] [PubMed] [Google Scholar]
- Frequency of aneuploidy in sperm from patients with extremely severe male factor infertility. Hum Reprod. 2005;20:2140-52.
- [CrossRef] [PubMed] [Google Scholar]
- Fluorescence in situ hybridisation sperm examination is significantly impaired in all categories of male infertility. Andrologia. 2018;50:e12847.
- [CrossRef] [PubMed] [Google Scholar]
- Male infertility: Establishing sperm aneuploidy thresholds in the laboratory. J Assist Reprod Genet. 2019;36:371-81.
- [CrossRef] [PubMed] [Google Scholar]
- Next-generation sequencing technology: Current trends and advancements. Biology (Basel). 2023;12:997.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative transcriptomic analysis of spermatozoa from high-and low-fertile crossbred bulls: Implications for fertility prediction. Front Cell Dev Biol. 2021;9:647717.
- [CrossRef] [PubMed] [Google Scholar]
- Mitochondrial functionality in male fertility: From spermatogenesis to fertilization. Antioxidants (Basel). 2021;10:98.
- [CrossRef] [PubMed] [Google Scholar]
- Exploring the ovine sperm transcriptome by RNAseq techniques. I Effect of seasonal conditions on transcripts abundance. PLoS One. 2022;17:e0264978.
- [CrossRef] [PubMed] [Google Scholar]
- Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422-32.
- [CrossRef] [PubMed] [Google Scholar]
- A role for chromosomal microarray testing in the workup of male infertility. J Mol Diagn. 2020;22:1189-98.
- [CrossRef] [PubMed] [Google Scholar]
- Seminal biomarkers for the evaluation of male infertility. Asian J Androl. 2016;18:426-33.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of genital tract markers in the human seminal plasma using an integrative genomics approach. Hum Reprod. 2013;28:199-209.
- [CrossRef] [PubMed] [Google Scholar]
- Non-invasive diagnostics of male spermatogenesis from seminal plasma: Seminal proteins. Diagnostics (Basel). 2023;13:2468.
- [CrossRef] [PubMed] [Google Scholar]
- Mitochondrial-induced epigenetic modifications: From biology to clinical translation. Curr Pharm Des. 2021;27:159-76.
- [CrossRef] [PubMed] [Google Scholar]
- Sperm and seminal plasma proteomics: Molecular changes associated with varicocele-mediated male infertility. World J Mens Health. 2020;38:472-83.
- [CrossRef] [PubMed] [Google Scholar]
- Mapping of seminal plasma proteins by two-dimensional gel electrophoresis in men with normal and impaired spermatogenesis. Mol Hum Reprod. 2001;7:715-22.
- [CrossRef] [PubMed] [Google Scholar]
- Proteomic analysis in seminal plasma of fertile donors and infertile patients with sperm DNA fragmentation. Int J Mol Sci. 2020;21:5046.
- [CrossRef] [PubMed] [Google Scholar]
- Basic principles and fundamental aspects of mass spectrometry In: Mass spectrometry in food analysis. United States: CRC Press; 2022. p. :3-17.
- [CrossRef] [Google Scholar]
- Proteome analysis using Gel-LC-MS/MS. Curr Protoc Protein Sci. 2019;96:e93.
- [CrossRef] [PubMed] [Google Scholar]
- LC-MS based detection of differential protein expression. J Proteomics Bioinform. 2009;2:416-38.
- [CrossRef] [PubMed] [Google Scholar]
- Cell-free circulating mitochondrial DNA: An emerging biomarker for airborne particulate matter associated with cardiovascular diseases. Free Radic Biol Med. 2023;195:103-20.
- [CrossRef] [PubMed] [Google Scholar]
- Urinary metabolome identifies signatures of oligozoospermic infertile men. Fertil Steril. 2014;102:44-53.e12.
- [CrossRef] [PubMed] [Google Scholar]
- Unravelling the power of OMICS for the infertile aging male. Curr Pharm Des. 2017;23:4451-69.
- [CrossRef] [PubMed] [Google Scholar]
- Untargeted GC-MS metabolomics. Methods Mol Biol. 2018;1738:133-47.
- [CrossRef] [PubMed] [Google Scholar]
- Metabolomic profiling of human spermatozoa in idiopathic asthenozoospermia patients using gas chromatography-mass spectrometry. Biomed Res Int. 2018;2018:8327506.
- [CrossRef] [PubMed] [Google Scholar]
- Metabonomic analysis of fatty acids in seminal plasma between healthy and asthenozoospermic men based on gas chromatography mass spectrometry. Andrologia. 2017;49:e12744.
- [CrossRef] [PubMed] [Google Scholar]
- Epigenetics: A key paradigm in reproductive health. Clin Exp Reprod Med. 2016;43:59-81.
- [CrossRef] [PubMed] [Google Scholar]
- Nanotechnology in reproductive medicine: Opportunities for clinical translation. Clin Exp Reprod Med. 2020;47:245-62.
- [CrossRef] [PubMed] [Google Scholar]







