Translate this page into:
Empowering of reproductive health of farm animals through genome editing technology

*Corresponding author: Dharmendra Kumar, Division of Animal Physiology and Reproduction, ICAR-Central Institute for Research on Buffaloes, Hisar, Haryana, India. dkumarbt@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Dua S, Bajwa KK, Prashar A, Bansal S, Beniwal M, Kumar P, et al. Empowering of reproductive health of farm animals through genome editing technology. J Reprod Healthc Med 2021;2:4.
Abstract
To cater the exponential growth of human population, need to improve food production and quality through modern biotechnology with limited recourses in a way that has minimal impact on the environment. The selective breeding and genomic selection have attended the momentum gain in livestock productivity. Recent advancement in genome-editing technologies offers exciting prospects for the production of healthy and prolific livestock. Genome editing involves altering genetic material by manipulation, addition, or removal of certain deoxyribonucleic acid (DNA) sequences at a specific locus in a way that does not occur naturally. The major genome editors are zinc finger nucleases, transcription-activator-like endonucleases, and clustered regularly interspaced short palindromic repeats associated protein nine systems which are proficient of cutting the nuclear DNA precisely at a predetermined position. This review provides an update on the use of genome editing systems to modify the genes related to reproduction of farm animal vis-à-vis human, update knowledge on the underlying mechanism and discusses new opportunities to produce genetically modified farm animals.
Keywords
Gene editing
Livestock
Clustered regularly interspaced short palindromic repeats-cas
Reproduction
Genetic modification
INTRODUCTION
Genome editing comprises altering genetic material by manipulation, either addition or removal of certain deoxyribonucleic acid (DNA) sequences at a specific locus in a way that does not occur naturally. Now, genome editing in livestock is instrumental for researchers to improve animal performance, efficiency and welfare, paving the way toward a more sustainable future of farm animals with the implementation of advanced biotechnologies. The number of genetically modified animals used in agriculture has increased significantly in recent years to meet the expected rise in demand for animal food products in the near future. Researchers have genetically engineered a number of mammals, from laboratory to farm animals including sheep,[1] goat,[2] pig,[3] and cow.[4] The purpose includes improvement in disease resistance, the better quality of milk and meat production which enhances the nutritional value of animal products. Earlier, the most accepted way of genetic modification was the direct manipulation of DNA sequences, which requires isolation, cut, and transfer specific DNA pieces to integrate into chromosome. Other techniques include manipulation of germ cells through in vitro fertilization, embryo transfer, cloning, and artificial insemination, which is actually more difficult, costly, time consuming, and bearing ethical issues. Recent advancements in genetic engineering (genome editing) enable us to perform genome level modifications precisely, as well as enhance the ease and speed with which farm animals could be genetically modified.[5,6] The genome editing tools facilitate targeted genetic alterations by creating site specific DNA double strand breaks (DSBs) at a preferred location in a genome. The DSBs in a genome can be repaired either by blunt-end, non-homologous end-joining (NHEJ) to introduce random mutations, or by the process of homologous DNA repair (HDR) to introduce a specific mutation by inserting an engineered fragment of DNA with homology on either side of the DNA break. Gene knockout or targeted insertion of a specific sequence in the genome could be obtained using proper DNA repair mechanism. In most cases, almost 90%, DSBs are repaired by the NHEJ mechanism which occurs during all stages of the cell cycle, with the two ends of the break being ligated, resulted in the formation of indels at the repair site. Zinc finger nucleases (ZFN) – the most common DNA binding domain found in eukaryotes, were the first genome editing nucleases observed in the genome editing technology.[7] It comprised ~ 30 amino acid modules which form two anti-parallel β-sheets opposing to α-helix, bind to approximately three specific base pairs in the major groove of the DNA. ZNFs have been designed in such a way to recognize all of the 64 possible trinucleotide combinations. Each ZFN typically recognizes 3–6 nucleotide triplets but it is functional only in the form of dimers; hence, pairs of ZFNs are required to target any specific locus: One that recognizes the upstream sequence and other recognizes the downstream sequence of the site to be modified. Transcription activator-like endonucleases (TALENS) – are similar to ZFNs, they use DNA binding motifs to direct the non-specific nuclease to cleave the genome at a specific site, but instead of recognizing triplets DNA, each domain recognizes a single nucleotide.[8,9] Clustered regularly interspaced short palindromic repeats (CRISPR)/ Cas (CRISPR-associated) – are ribonucleic acid (RNA)-based bacterial defense mechanisms designed to recognize and eliminate foreign DNA from invading bacteriophage and plasmids.[10] They contain Cas endonuclease that is directed to cleave a target sequence by guide RNA (gRNA).[11] Targeted genetic alterations induced by these genome editors, enhances the DNA mutation rate through induction of double-strand breaks at a predetermined genomic site. Compared to conventional homologous recombination based gene targeting, genome editors increase the targeting rate ~10,000-fold through gene disruption and subsequent mutagenic DNA repair also stimulated at a similar frequency.[12]
Among the above-mentioned three genome editors, CRISPR/ Cas9 now appears as superior over ZFNs and TALENs due to its multiple genomic sites targeting capability. Comparisons of these genome editors are enumerated in [Table 1]. Therefore, the present review provides an update on the use of CRISPR/Cas9 to modify the genes related to reproduction of farm animal’s vis-à-vis human, update knowledge on the underlying mechanism, and discusses new opportunities to produce genetically modified farm animals.
| Features of gene editing tools | ZFNs | TALENs | CRISPR/CAS 9 |
|---|---|---|---|
| Target DNA recognition | Protein-DNA | Protein-DNA | RNA-DNA |
| Methylation sensitivity | Sensitive | Sensitive | No |
| Type of nuclease | ZF-Fok1 fusion protein | TALE-Fok1 fusion protein | Guide RNA and CAS 9 Protein |
| Construct | Zinc Finger sequence specifically recognize 3 bp linked to Fok 1 | Protein sequence specific to binding a nucleotide sequence (1:1) linked to FoK1 nuclease | A 20 nt crRNA fused to tracrRNA and Cas 9 endonuclease |
| Recognition site | Typically, 18–36 bp (usually a multiple of 3) per ZFN pair | Typically, 28–40 bp per TALEN pair | 22 bp followed immediately by 5’- NGG–3’ PAM |
| Targeting specificity | Small number of positional mismatches tolerated | Small number of positional mismatches tolerated | Positional and multiple consecutive mismatches tolerated, but it is easier to predict possible sites using Watson-Crick base pairing rules |
| Target constraints | Difficult to target Non guanine rich regions | Requires offset binding proteins with defined spacing | Targeted sequence must immediately precede aa PAM site |
| Mechanism of action | ZF proteins recognize target DNA sequences dimerization of Fok I nucleases induces DSBs of DNA. DSBs are repaired by NHEJ or HDR | TALE proteins recognize target DNA sequences dimerization of Fok I nucleases induces DSBs of DNA. DSBs are repaired by NHEJ or HDR | Guide RNA recognizes target DNA sequence next to a NGG motif Cas9 induces DSBs of DNA. DSBs are repaired by NHEJ or HDR |
| Time for vector construction | Months | 2–4 weeks | 1–2 weeks |
| Ease of design/engineering | Difficult: may require substantial protein engineering | Moderate: Requires complex molecular cloning, but TALEn design has been streamlined by the availability of modules of repeat combinations that reduce the amount of clothing | Easy: Simple to design and guide sequences are easily engineered using standard cloning procedures or oligo synthesis |
| Multiplexing ability advantages | No Small protein size (< 1 kb) | No High specificity | YES Enables multiplexing (Targeting multiple genes) |
| Advantages | Highly efficient and specific | Highly efficient and specific | Highly efficient, easy to be constructed and capable of editing multiple sites simultaneously |
| Disadvantages | Large-scale screening (Length of target sequence confined to multiples of 3), time-consuming, and expensive to be constructed | Tedious and time consuming to be constructed | PAM motif next to target sequence required Cas9 nuclease is large (4.2 kb) |
ZFN: Zinc finger nucleases, CRISPR: Clustered regularly interspaced short palindromic repeats, TALEN: Transcription activator-like endonucleases, RNA: Ribonucleic acid, DNA: Deoxyribonucleic acid, PAM: Protospacer adjacent motif, NHEJ: Non-homologous end joining, HDR: Homologous DNA repair, DBAs: Double strand breaks
CRISPR/CAS9 MEDIATED GENE EDITING IN FARM ANIMALS
Genome editing in farm animals has tremendous practical applications, such as a large animal could be a more accurate model for medical research due to its anatomical and physiological similarities and through improved animal welfare that helping to secure a sustainable future for livestock farming. Genetically engineered large animal models could play a decisive role in the study of complex pathophysiological and physiological processes, and introducing new traits of commercial value. Using genomic selection more precisely predicts the potential of animals at an early age than the classical pedigree index.[13] The classical approaches to produce transgenic mice through pronuclear injection, intracytoplasmic injection, and embryonic stem cell-mediated genetic modification are time consuming and expensive, and most importantly are not well suited to other large mammalian species. Sometimes, it can take more than a decade to slightly improve the percentage of lean meat using traditional breeding programs. These classical techniques are currently replaced by new and promising technology, CRISPR/Cas9, and are successfully translated to other species including livestock.[14,15]
The CRISPR-Cas system naturally exists in the prokaryotes as an adaptive immunity mechanism that enables to detect and protect against mobile genetic elements. The system exists across diverse species of bacteria and archaea, which differ in their cell components and action. Overall there are six CRISPR-cas types and 29 subtypes available currently, and this list is undergoing rapid expansion. The system contains CRISPR binds with a gRNA for guidance and target specificity, and an associated endonuclease Cas9 for cleavage. Binding specificity is thus dependent on RNA-DNA interaction strength. The cas9 along with a gRNA locate a specific sequence using proto-spacer adjacent motifs (PAMs) and then splice foreign DNA at precise locations. To make the functional DNA-target complex, Cas9 needs two distinct RNA transcripts, CRISPR RNA (crRNA) and trance-acting CRISPR RNA (tracrRNA). Finally, single-gRNA (sgRNA) complement to the target sequence which is adequate to program Cas9 to create a DSB in target DNA.[16,17] The DSB is thereafter repaired by various endogenous mechanisms including NHEJ, HDR, homology independent target integration (HITI), and single base editing. When CRISPR makes the site-specific DSB, DNA repair mechanisms are employed to repair the break [Figure 1]. If the complementary sequence of DNA is available at the cleavage site, HR could be operational to repair the damage; otherwise, the cell will use NHEJ.[11] NHEJ causes deletion or insertion of a small number of nucleotides, which can be harnessed to create gene knock-out. Recently in HITI, a tag that is flanked with gRNA target sites issued to release Cas9 from a plasmid and cleave a recipient genomic target adjacent to the gene of interest. Recently, an HDR-dependent strategy termed “CORRECT” (consecutive re-guide or re-Cas steps to erase CRISPR-Cas blocked targets) has been developed to produce scar free targeted knock-in of generation of disease-relevant mutations.[18]
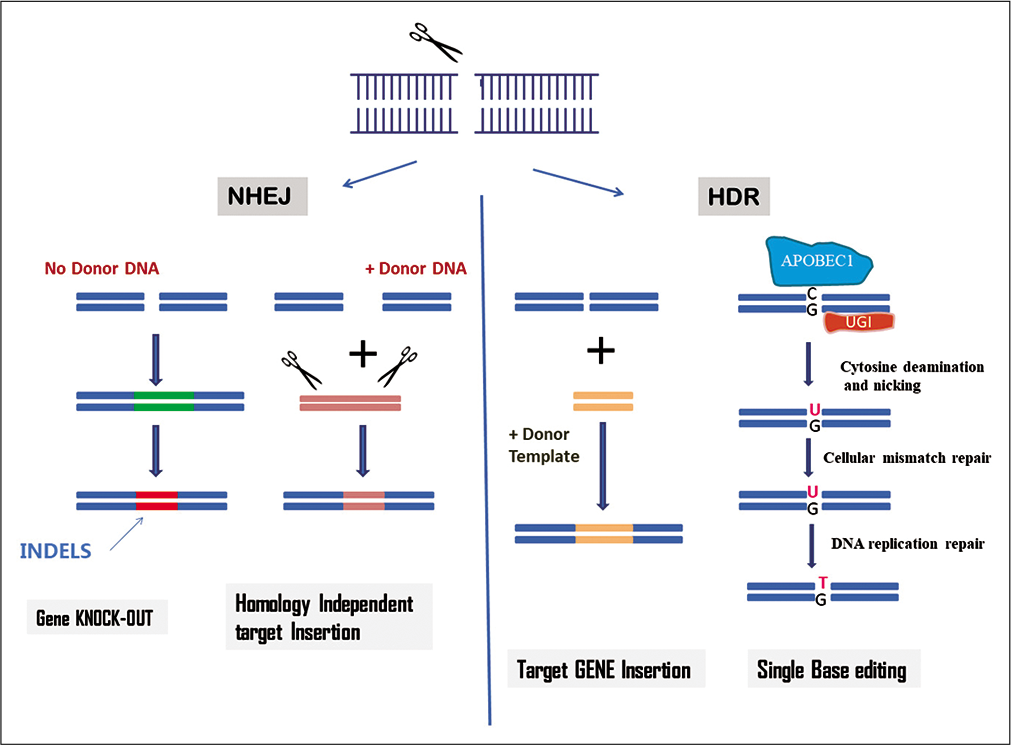
- Mechanism of clustered regularly interspaced short palindromic repeats-Cas9 system for genome editing.
Gene editor has the ability to edit a single nucleotide base which is important for the construction of genetic disease models and the development of corrective therapeutics as mostly genetic variants of human disease is related to point mutation. Targeted HDR-dependent DSB repair remains ineffective, particularly in post-mitotic cells with reduced HDR activity and probabilities to induce off-target mutagenesis, and even on-target activation can adversely affect cell viability.[19] Recently, site-specific single base editing tools without triggering DSB have been developed to create animal models with nonsense mutation or single amino acid substitution. These are cas9 nickase also known as dcas9 (deficient cas 9) and a third generation editor BE3. For a single-nucleotide C→T (or G→A) conversion, Cas9 nickase has been fused to cytidine deaminases such as apolipoprotein B mRNA-editing enzyme, can achieve targeted nucleotide conversion within a 5-bp activity window located in a spacer sequence. Lately, the base editor toolbox has been extended to adenine base editors, which can perform targeted A→G (or T→C) nucleotide conversions.[20]
Although, CRISPR cas system has been known to manipulate DNA, recent advancement is carried out to develop CRISPRCas technology for binding and cleaving specific RNA to achieve RNA targeting applications. Rcas9, (RNA targeting Cas9) system, use PAMers that recognizes and provided to direct cas9 to target RNA sequences, which lack PAMs at the corresponding genomic DNA site. Rcas9 could be used to control cellular processes at transcript level because it can stimulate site-specific cleavage of ssRNA.[21]
USE OF GENOME EDITING FOR EMPOWERING REPRODUCTION OF FARM ANIMALS
The application of CRISPR-Cas in livestock as reproductive technologies in which efficient delivery of the CRISPR system into zygotes or reproductive cells would able to change the individual hereditary genetic profile; thus be transmitted to the future generations.[22] Reproduction is one of the best suited biological systems in which gene manipulation such as knockout can be successfully applied [Figure 2]. Conventional techniques such as somatic cell nuclear transfer and zygote microinjection are technically challenging, labor-intensive, costly, and require expert skills with bulky micromanipulation equipment; restricting their use to a small number of specialized laboratories[23] Recent advancement in genome editing technique simplifies the delivery of CRISPR-Cas system into livestock reproductive cells such as zygote electroporation or transduction with recombinant adeno-associated virus. The novel surrogate sire technology (SST) is a new toolkit for delivering CRISPRCas to reproductive cells. The straightforwardness of these methodologies will allow performing genome editing in farm animals on-site. Given the availability of large animal genome sequences and the advancement of nuclease-mediated gene editing technologies, the efficient modification of elite phenotypes in cattle,[24,25] sheep, pig,[3,26-30] dog,[31] and goat[2] developed in the past few years is shown in [Table 2].
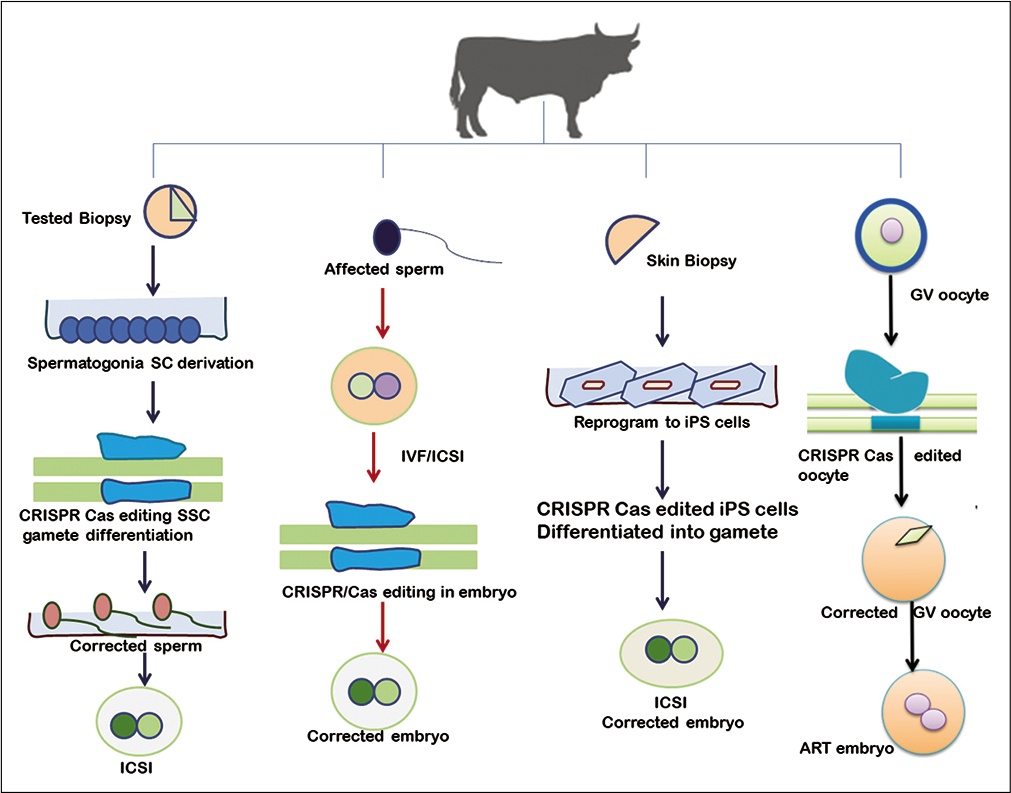
- Diagrammatic illustration of clustered regularly interspaced short palindromic repeats/Cas9 application in reproductive cell gene editing.
| Animal species | Cell type | Approach | Gene target and function | Modification | Reference |
|---|---|---|---|---|---|
| Pig | Porcine fetal fibroblast | Random mutagenesis | β–glucanase, xylanase, phytase gene to improve, pig feed efficiency | Transgenic | 47 |
| Porcine fetal fibroblast | ZFN | MSTN to improve meat quality and production | NHEJ-KO | 48 | |
| Porcine fetal fibroblast | CRISPR/Cas | Exon of CD 163 replaced by CD163L1 of human | HDR KI-KO | 3 | |
| Zygote | ZFN | PPAR–γ disease model for metabolic disease | NHEJ-KO | 49 | |
| Zygote | TALEN | LDLR, disease model for hyper cholesterolemia | NHEJ-KO | 50 | |
| Zygote | CRISPR/Cas | NANOS 2 | NHEJ-KO | 25 | |
| Cattle | Bovine fetal fibroblast | ZFN | BLG Cattle milk production | NHEJ-KO | 51 |
| Skin fibroblast | CRISPR/Cas | Repaired IARS syndrome, mutation in IARS gene | HDR (single base substitution) | 52 | |
| Goat | Zygote | CRISPR/Cas | MSTN target gene | NHEJ-KO | 2 |
| Dog | Zygote | CRISPR/Cas | ApoE, Disease model for cardiovascular disease | NHEJ-KO | 53 |
ZFN: Zinc finger nucleases, CRISPR: Clustered regularly interspaced short palindromic repeats, TALEN: Transcription activator-like endonucleases, PPAR–γ: Peroxisome proliferator-activated receptor gamma, LDLR: Low density lipoprotein receptor, BLG: Beta-lactoglobulin, IARS: Isoleucyl-tRNA synthetase, NHEJ: Non-homologous end joining, HDR: Homologous DNA repair
GENE EDITING IN ZYGOTE OF FARM ANIMALS
Zygote electroporation is a newly developed technique that allows the direct introduction of CRISPR-Cas into zygotes which are suspended in a suitable medium by application of voltage. During electroporation, the electric pulses create pores in the membrane of the zygote, which allow CRISPRCas to pass through the pores and enter into the nucleus where CRISPR-Cas integrates into a DNA sequence and initiates editing activity.[32] However, optimizations of pulse duration and voltage strength require attention to maximize transfection efficiency while maintaining embryo viability. Another technique such as zona-intact zygote electroporation has now employed to successfully edit the pigs and cattle genomes.[26] Namula et al. have successfully produced biallelic MSTN gene mutant in bovine blastocyst by electroporation of cas9 into the viable embryo at 10 h after the start of insemination.[33]
GENE EDITING OF MALE REPRODUCTIVE CELLS
An alternative to zygote/embryo editing could be achieved through targeting sperm or oocyte or even primordial germ cells. The edited sperms and oocytes can be used in artificial reproduction techniques to produce healthy embryos. The spermatogenic or haploid germ cell genes that are important for fertility are more specific to the gonads and often comprise a single exon.[34] The breeding system is associated with creating homozygous genetic manipulations in genes involved in gametogenesis, fertilization, and conception. Gene disruption methods have long been advantageous to study male reproductive biology. The CRISPR/Cas9 technology can be used to generate gene disrupted mouse models to investigate the genetics of male fertility that look at the essential genetic component of male reproductive system.[35] With ease and precision of CRISPR/Cas9 system this has revolved around with worth pursuing ability to produce fertility or infertility phenotype within few months, in addition to the introduction of point mutation, ability to further investigate the molecular and epigenetic mechanisms of fertility related genes.[36] CRISPR/Cas9 system could be used in immature sperm cells to produce gene-corrected mature sperm cells, which further improve the outcome of assisted reproduction. In male germline cells, the mature post-meiotic sperms could not be subjected to genetic manipulations; however, spermatogonial stem cells (SSCs) seem a better option for manipulation, particularly for the infertility patients having no sperm cells due to maturation arrest. SSCs can be propagating efficiently in culture and can be transplanted back into the testis to generate mature and functional sperm.[37] Alternative technology named SST outlined to generate male animals without endemic germline stem cells, act as a surrogate sire. When SSCs from a donor male transplanted into recipient testes of a surrogate sire, providing a source of self-propagating stem cells which can produce mature sperm containing the genetic information of the donor.[37] The recipient male can then disseminate the same genetic information of the donor by natural breeding or through artificial insemination system [Figure 2].[38]
In a study, the crygc gene in SSCs is mutated through CRISPR/Cas9 system, which further undergone spermatogenesis after transplantation into seminiferous tubules of infertile mouse testes. Moreover, CRISPR/ Cas9 induced disease-causing mutations in crygc gene of SSCs is repaired by NHEJ in mouse, resulting in SSCs carrying modified gene with no off-target mutations at entire genome sequencing and the offspring showed 100% efficient phenotype.[39] CRISPR/Cas9 catalyzed the rat SSCs to regenerate spermatogenesis and showed long-term sperm-producing capability following transplantation into rat testes. Targeted mutations in germ line cell genes such as epithelial stromal interaction 1 (Epsti1) and receptor tyrosine protein kinase 3 (Erbb3) were vertically transmitted from recipient to exclusively produce non-mosaic offspring. Monoclonal enrichment of Erbb3null germlines exposed recessive spermatogenesis defects and yielded modified offspring isogenic at targeted alleles. Therefore, spermatogonial gene modification with CRISPR/ Cas9 provided a platform to produce targeted germ line modifications in rats and to study spermatogenesis.[38]
Recently, knockout mouse models for testis specific genes (Slx2) were generated to study the spermatogenesis process in male reproductive systems in vivo. The study revealed that slx2 itself does not play an essential role in spermatogenesis but CRISPR/Cas9 technology could help to boost up the functional study of X-linked genes of testis in vivo.[40] More recently, male pigs have been produced with ablated germ line by editing of the NANOS2 gene using CRISPR/Cas9 showed novel applications for germ cell transplantation (surrogate sires) for accelerating genetic selection of male animals.[41] The generated animals are physiologically healthy; displaying no abnormal phenotypes other than the complete lack of native SSCs. The work is under progress to knockout NANOS2 gene for the generation of surrogate sire in cattle, sheep, and goats. In the future, it could be possible to predict the elite SSCs from sexually immature males using genomic tools, could obtain SSCs from that male and transplanted into ablated germline recipients to obtain functional sperm within months. These approaches will reduce the waiting period until breeding maturity for each animal, generation intervals could be escaped, thus accelerating the rate of genetic gain.
GENE EDITING OF FEMALE REPRODUCTIVE CELLS
An alternative to zygote editing, genome modifications could also be applied during gametogenesis. An oocyte is more easily accessible for gene targeting than zygote but technical hurdle remains such as scarce availability of oocytes and high sensitivity toward electrical pulse number. In this aspect, the CRISPR/Cas9 system could be applied to the growing oocytes to generate genetically modified oocytes, which would be subsequently useful for assisted reproductive technology, in overcoming the off-target mutations in the following generation.[22] The establishment of the gene editing by electroporation of the Cas9 protein (GEEP) method to generate high-efficiency disruption of the targeted gene, overcome the complexity associated with microinjection of the CRISPR/Cas9 system into Zygote [Figure 2]. The GEEP method is a simple method for the transduction of CRISPR/ Cas9 which requires the introduction of Cas9 protein and sgRNA into either matured oocytes or in vitro fertilized zygotes by electroporation. Many studies have been carried out using the GEEP technique in farm animal species such as pigs[42] and bovine.[33] Recently, it was demonstrated that mutations created in oocyte gene of TGF-beta growth factors family and its related receptor, growth differentiation factor 9, bone morphogenetic protein 15, and activin receptor-like kinase 6 resulted in increased ovulation rates and thus litter size.[43]
Recently in humans, one oocyte nucleus and two sperm nuclei (tri-pronuclear zygotes) were generated with a cleaved endogenous β-globin gene which later on repaired using CRISPR/Cas9 through the non-crossover HDR mechanism.[44] Another group edited embryos directly through cytoplasmic pronuclei microinjection of zygote with no off-target genetic modifications. Although the efficiency of genome editing in embryos is low, still several studies have been done in different animals such as rat, sheep, cattle, pigs, and dogs to demonstrate the feasibility and efficiency of genome editing in animals.[45,46] This technique can prevent the chances of genetic disorders, for example, cataract development demonstrated successful 1 bp deletion in a specific gene in mouse offspring.[47] Microinjection of Cas9 or TALENs in non-human primates led to the birth of modified offspring.[48] The efficiency of genetic modifications by Cas9 or TALENs in mammalian zygote ranges from 0.5% to 40.9% per injected zygote. Recently, CRISPR/Cas9 editing technique was performed in the human zygote to validate its specificity and fidelity.[49] The edited embryos were mosaic which is similar to the findings in other model systems. Furthermore, off-target mutations were observed by CRISPR-Cas9 complex acting in another part of the genome. Notably, a specific portion of the genome known as exome was confirmed for off-target mutations and such mutations could be higher due to the mosaic nature of gene correction in edited embryos.[22]
In conclusion, current genome editing technologies particularly CRISPR/Cas9 is not only precise but also inexpensive and very efficient. This potent technology allows animal breeders to precisely and efficiently edit genome either to introduce desirable traits, such as hornless, heat tolerance, and disease resistance or to remove deleterious mutations, such as recessive lethal genetic variants; without affecting other genetic characteristics of their herd.[50-53] Further, there is increased acceptance and use of techniques for assisted reproduction, which is also likely to be required for genome editing in human embryos.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- CRISPR/Cas9-mediated sheep MSTN gene knockout and promote sSMSCs differentiation. J Cell Biochem. 2019;120:1794-806.
- [CrossRef] [PubMed] [Google Scholar]
- Use of CRISPR/Cas9 technology efficiently targetted goat myostatin through zygotes microinjection resulting in double-muscled phenotype in goats. Biosci Rep. 2018;38:BSR20180742.
- [CrossRef] [PubMed] [Google Scholar]
- Generation of pigs resistant to highly pathogenic-porcine reproductive and respiratory syndrome virus through gene editing of CD163. Int J Biol Sci. 2019;15:481-92.
- [CrossRef] [PubMed] [Google Scholar]
- Single Cas9 nickase induced generation of NRAMP1 knockin cattle with reduced off-target effects. Genome Biol. 2017;18:13.
- [CrossRef] [PubMed] [Google Scholar]
- Basics of genome editing technology and its application in livestock species. Reprod Domest Anim. 2017;52:4-13.
- [CrossRef] [PubMed] [Google Scholar]
- Application of genome editing in farm animals In: Genomics and Biotechnological Advances in Veterinary, Poultry, and Fisheries. Cambridge: Academic Press; 2019. p. :131-49.
- [CrossRef] [Google Scholar]
- An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778-85.
- [CrossRef] [PubMed] [Google Scholar]
- Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757-61.
- [CrossRef] [PubMed] [Google Scholar]
- A tale nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143-8.
- [CrossRef] [PubMed] [Google Scholar]
- Evolution and classification of the CRISPRCas systems. Nat Rev Microbiol. 2011;9:467-77.
- [CrossRef] [PubMed] [Google Scholar]
- ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397-405.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular scissors and their application in genetically modified farm animals. Transgenic Res. 2015;24:381-96.
- [CrossRef] [PubMed] [Google Scholar]
- In vitro breeding: Application of embryonic stem cells to animal production. Biol Reprod. 2019;100:885-95.
- [CrossRef] [PubMed] [Google Scholar]
- Genome editing in livestock: Are we ready for a revolution in animal breeding industry? Transgenic Res. 2017;26:715-26.
- [CrossRef] [PubMed] [Google Scholar]
- Genome editing approaches to augment livestock breeding programs. J Exp Biol. 2020;223:jeb207159.
- [CrossRef] [PubMed] [Google Scholar]
- A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;37:816-21.
- [CrossRef] [PubMed] [Google Scholar]
- The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20:490-507.
- [CrossRef] [PubMed] [Google Scholar]
- CRISPR-CAs9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24:927-30.
- [CrossRef] [PubMed] [Google Scholar]
- Programmable base editing of A-T to G-C in genomic DNA without DNA cleavage. Nature. 2017;551:464-71.
- [CrossRef] [PubMed] [Google Scholar]
- CRISPR RNA-dependent binding and cleavage of endogenous RNAs by the Campylobacter jejuni Cas9. Mol Cell. 2018;69:893-905.
- [CrossRef] [PubMed] [Google Scholar]
- Genome engineering through CRISPR/Cas9 technology in the human germline and pluripotent stem cells. Hum Reprod Update. 2016;22:411-9.
- [CrossRef] [PubMed] [Google Scholar]
- Somatic cell nuclear transfer followed by CRIPSR/ Cas9 microinjection results in highly efficient genome editing in cloned pigs. Int J Mol Sci. 2016;17:2031.
- [CrossRef] [PubMed] [Google Scholar]
- Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res. 2011;21:1638-40.
- [CrossRef] [PubMed] [Google Scholar]
- Establishment of protocol for preparation of gene-edited bovine ear-derived fibroblasts for somatic cell nuclear transplantation. Biomed Res. 2018;39:95-104.
- [CrossRef] [PubMed] [Google Scholar]
- Simplified pipelines for genetic engineering of mammalian embryos by CRISPR-Cas9 electroporation. Biol Reprod. 2019;101:177-87.
- [CrossRef] [PubMed] [Google Scholar]
- Novel transgenic pigs with enhanced growth and reduced environmental impact. Elife. 2018;7:e34286.
- [CrossRef] [PubMed] [Google Scholar]
- Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci Rep. 2015;5:14435.
- [CrossRef] [PubMed] [Google Scholar]
- Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011;21:979-82.
- [CrossRef] [PubMed] [Google Scholar]
- Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109:17382-7.
- [CrossRef] [PubMed] [Google Scholar]
- Generation of ApoE deficient dogs via combination of embryo injection of CRISPR/Cas9 with somatic cell nuclear transfer. J Genet Genomics. 2018;45:47-50.
- [CrossRef] [PubMed] [Google Scholar]
- Genome editing in large animals: Current status and future prospects. Natl Sci Rev. 2019;6:402-20.
- [CrossRef] [Google Scholar]
- Genome mutation after the introduction of the gene editing by electroporation of Cas9 protein (GEEP) system into bovine putative zygotes. In Vitro Cell Dev Biol Anim. 2019;55:237-42.
- [CrossRef] [PubMed] [Google Scholar]
- Fertilization: A sperm's journey to and interaction with the oocyte. J Clin Invest. 2010;120:984-94.
- [CrossRef] [PubMed] [Google Scholar]
- Advantages of using the CRISPR/Cas9 system of genome editing to investigate male reproductive mechanisms using mouse models. Asian J Androl. 2015;17:623-7.
- [CrossRef] [PubMed] [Google Scholar]
- Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3:3355.
- [CrossRef] [PubMed] [Google Scholar]
- Spermatogonial stem cell transplantation: Insights and outlook for domestic animals. Annu Rev Anim Biosci. 2019;7:385-401.
- [CrossRef] [PubMed] [Google Scholar]
- On-farm livestock genome editing using cutting edge reproductive technologies. Front Sustain Food Syst. 2019;3:1-8.
- [CrossRef] [Google Scholar]
- Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25:67-79.
- [CrossRef] [PubMed] [Google Scholar]
- CRISPR/Cas9 promotes functional study of testis specific X-linked gene in vivo. PLoS One. 2015;10:e0143148.
- [CrossRef] [PubMed] [Google Scholar]
- Generation of germline ablated male pigs by CRISPR/Cas9 editing of the NANOS2 gene. Sci Rep. 2017;7:40176.
- [CrossRef] [PubMed] [Google Scholar]
- Genome mutation after introduction of the gene editing by electroporation of Cas9 protein (GEEP) system in matured oocytes and putative zygotes. In Vitro Cell Dev Biol Anim. 2019;55:237-42.
- [CrossRef] [PubMed] [Google Scholar]
- Oocyte-expressed genes affecting ovulation rate. Mol Cell Endocrinol. 2005;234:57-66.
- [CrossRef] [PubMed] [Google Scholar]
- CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363-72.
- [CrossRef] [PubMed] [Google Scholar]
- Generation of gene-target dogs using CRISPR/Cas9 system. J Mol Cell Biol. 2015;7:580-3.
- [CrossRef] [PubMed] [Google Scholar]
- Knock-in fibroblasts and transgenic blastocysts for expression of human FGF2 in the bovine β-casein gene locus using CRISPR/Cas9 nuclease-mediated homologous recombination. Zygote. 2016;24:442-56.
- [CrossRef] [PubMed] [Google Scholar]
- Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659-62.
- [CrossRef] [PubMed] [Google Scholar]
- Generation of gene-modified cynomolgus monkey via Cas9/ RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836-43.
- [CrossRef] [PubMed] [Google Scholar]
- Genome editing in human embryos: Ethical concerns and practical applications. Jpn J Clin Hematol. 2019;60:1033-45.
- [Google Scholar]
- Breeding and genetics symposium: Breeding heat tolerant dairy cattle: The case for introgression of the slick prolactin receptor variant into dairy breeds. J Anim Sci. 2017;4:1788-800.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of gene editing versus conventional breeding to introgress the POLLED allele into the US dairy cattle population. J Dairy Sci. 2019;5:4215-26.
- [CrossRef] [PubMed] [Google Scholar]
- Genome editing for disease resistance in pigs and chickens. Anim Front. 2019;9:6-12.
- [CrossRef] [PubMed] [Google Scholar]
- CRISPR in livestock: From editing to printing. Theriogenology. 2020;150:247-54.
- [CrossRef] [PubMed] [Google Scholar]






