Translate this page into:
Biomarkers in ovarian cancer and saliva: An update

*Corresponding author: Savita Yadav, Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India. savita11@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Chandra KB, Yadav S. Biomarkers in ovarian cancer and saliva: An update. J Reprod Healthc Med 2021;2:1.
Abstract
Asymptomatic nature at the initial stage and heterogeneity makes ovarian cancer a “silent killer” which is being considered as most lethal gynecological cancer by acquiring the fifth leading cause of cancer-related deaths in women. Detection of ovarian cancer frequently requires painful invasive procedures such as multiple biopsies and blood tests which results in an undue stress and discomforts in patient. Recently, saliva is being opted as an alternative source for biomarker discovery due to procedure being non-invasive collection method cost-effectiveness, ease of sample collection, and handling. Saliva, a multiconstituent oral fluid secretion from the major and minor salivary glands enriched with proteins, DNAs, RNAs, and metabolites, behaves as a “mirror of the body.” Salivary diagnostics has become booming field with the development of new and advanced technologies accompanying with proteomics, RNA sequencing, liquid biopsy, and point-of-care (POC) diagnostics. Reliable and reproducible biomarkers identified through advanced salivaomics techniques such as proteomics, transcriptomics, genomics, and metabolomics for oral and systemic diseases including cancers can serve as a diagnostic and monitoring tool. Scientific communities are engaged in developing new technologies for the identification and validation of an extensive range of salivary biomarkers that will provide clinical and scientific credibility for saliva. This review provides a comprehensive update about the significant salivary biomarkers identified by the omics method that can be used for the early detection of ovarian cancer. In coming years, salivaomics may become a very important tool for early detection of ovarian cancer and salivary biomarkers may translate into improvement of treatment outcome and increased survival rate.
Keywords
Genomics
Ovarian cancer diagnosis
Proteomics
Saliva
Salivary biomarkers
SIGNIFICANCE OF SALIVA
Human saliva is being opted as an alternative source for biomarker discovery due to non-invasive procedure, cost-effectiveness, easy to handle, and undemanding collection process over blood serum or plasma. It is a clear, colorless, and odorless hypotonic solution of pH 6.0–7.0 secreted from three major salivary glands, that is, parotid, submandibular, and sublingual glands along with 300–400 minor salivary glands (approximately) in the oral cavity [Figure 1].[1] The autonomous nervous system efficiently regulates the flow rate and composition of saliva which depends on the signaling by neuropeptides and intracellular calcium.[2] Almost 1–1.5 L of saliva is being produced on daily basis by a healthy individual consisting of 99% of water, 0.2% inorganic substances, and 0.3% proteins with a flow rate of 0.3–0.7 ml saliva/min.[3,4] The heterogeneous biofluid plays several important roles in facilitating tissue lubrication, swallowing, tasting, and digestion, also behaves as an antibacterial and antifungal agent by creating a protective barrier against pathogens.
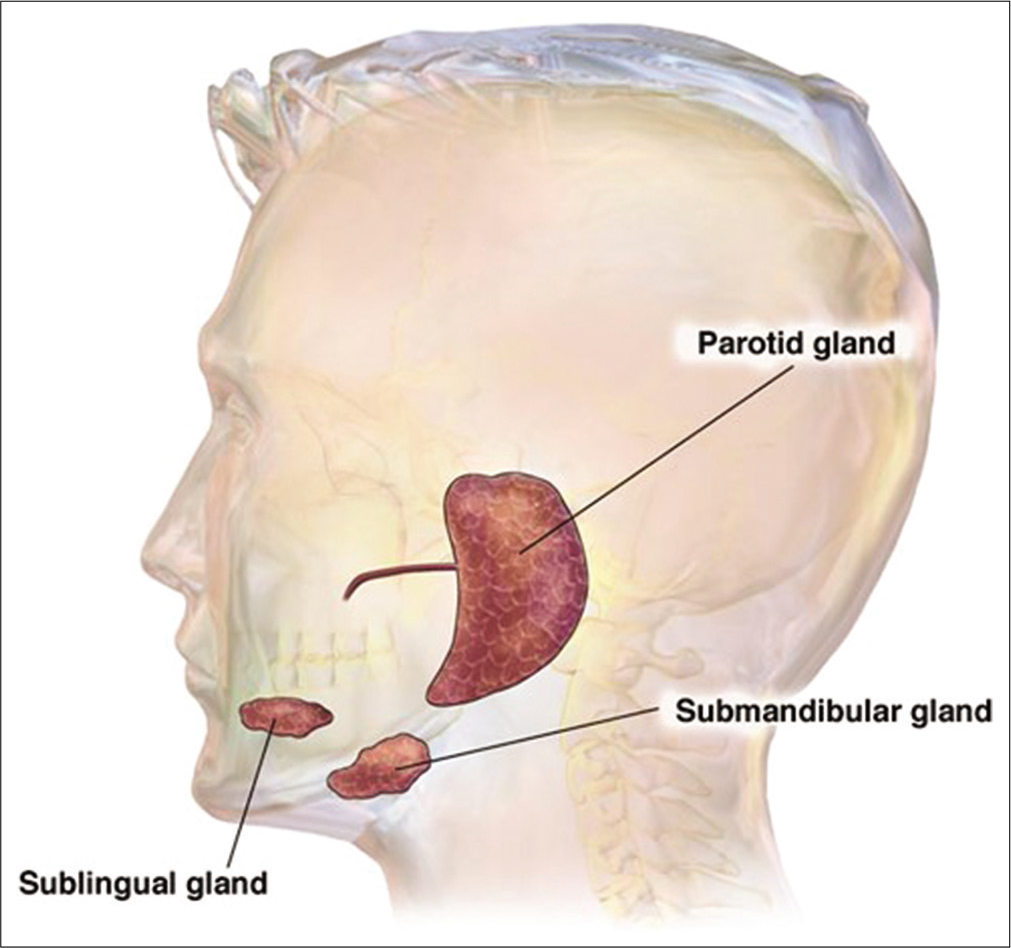
- Location of three major salivary glands parotid, submandibular, and sublingual glands. The parotid glands are the largest of the three major salivary glands and located just in front of each ear. The submandibular glands are just beneath the lower jaw just posterior and below, the sublingual glands are located under the tongue. Each gland is in pair. (Image courtesy – Image has been taken by GOOGLE from the site of Studio Dentaire Montréal).
Like blood, the multiconstituent oral biological fluid contains several molecular and cellular entities that are capable of communicating the current status of an individual’s health and often called “a window on health status” or “a mirror of the body.”[4] An extensive range of proteins, mRNA, microRNA (miRNA) transcripts, genomes, metabolites, and microbiota present in saliva is involved in various biological events associated with the disease onset and progression.[5] Different constituents of saliva are subject to comprehensive evaluations and their composition changes under different pathological conditions.[6] Any altered expression in the concentration, function, or structure can be linked with the pathogenesis.[7] During pathogenesis, overexpression of any protein can be cleaved from cancer cells surface and shed directly into saliva. Saliva and blood are two different biofluids that may be linked to each other on the molecular level and molecules from blood also reach into saliva through various mechanisms such a ultrafiltration and transdiffusion.[8,9]
Previously, it was assumed that an extracellular fluid from the oral cavity can be used only for the detection of oral diseases. However, nowadays, saliva is widely used for liquid biopsy for various systemic disorders including cancer due to the presence of an ample number of potential biomarker candidates.[4,10] Liquid biopsy is a revolutionary technique for identification and isolation of genomic biomarkers in cancer patients which will help to establish a more effective modified therapeutic algorithms and real-time therapy monitoring of disease.[4,10] We are exploring saliva as an alternative potential source for diagnosis over to blood and tissue due to superabundance of potential biomarkers. Identification and application of saliva-based biomarkers make saliva a precious means for diagnosis of different systemic diseases including cancer.
SALIVARY PROTEOMICS
Saliva is achieving a status of the plentiful source of biomarkers for oral and systemic diseases due to the presence of a large array of molecules. Scientific communities are wholeheartedly welcoming saliva as a unique and informative biological fluid for both research purpose and clinical diagnosis due to its non-invasive and cost-effective collection process. The study of saliva and constituents through recent and advanced techniques such as proteomics, transcriptomics, genomics, and metabolomics is “Salivaomics.” Various reliable and reproducible salivary biomarkers have been identified through these advanced techniques for oral and systemic diseases[11,12] which reflect the physiological and pathological condition of patients and can be used for diagnosis, prognosis, and drug monitoring and pharmacogenetic studies.[13,14] Approximately 40% of biomarkers can be identified from saliva by advanced and effective diagnostic alphabets for various systematic diseases including cancer [Figure 2].[15]
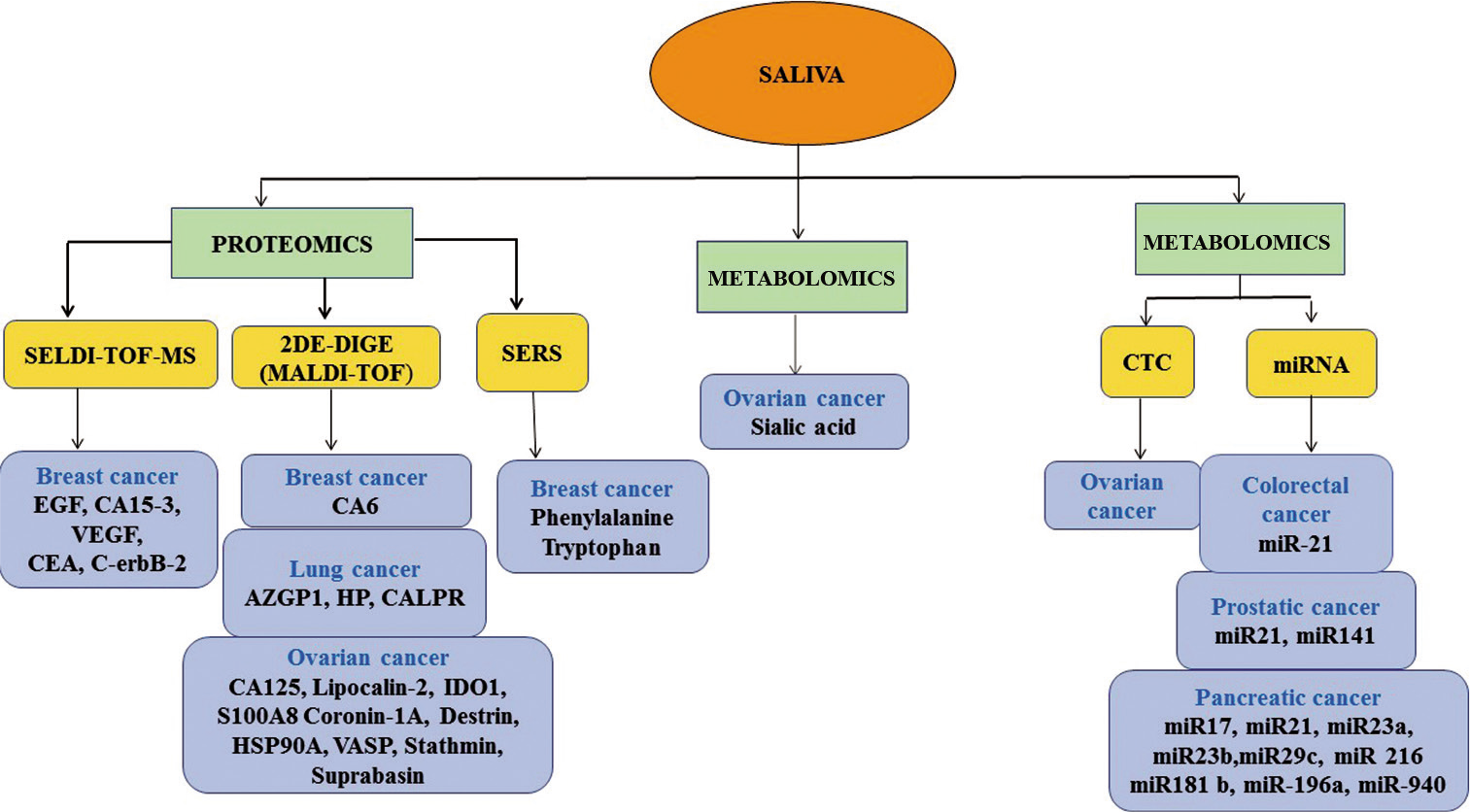
- Biomarkers identified from saliva by recent and advanced technologies for various malignancies.
Proteins present in saliva involved either in pathogenesis or therapeutics can be explored by proteomics; a technique used to analyze the protein composition, interactions, and altered expressions qualitatively and quantitatively.[16,17] Saliva is a diluted and less complex biofluid having total protein concentration 0.7–2.4 mg/ml[18] and shares 30% of the proteome with the blood or plasma proteome.[15,19] Salivary proteomics is regarded as a novel approach for discovery, verification, and validation of protein[20] and salivary proteome is highly inclined toward biochemical and physiological processes. Understanding the role of salivary proteome will be required to gain insight into physiological and pathological processes relevant for the identification of biomarkers.[21] Two-dimensional gel electrophoresis combined with mass spectrometry and bioinformatics tools for data acquisition and management helps the basic scientist to analyze salivary proteomes.[22] At present, salivary proteomics is being popular for identification and validation for potential biomarkers by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS).[23] Recently, near about 3000 differentially expressed proteins and peptides were identified from saliva which were post-translationally modified with glycosylation, proteolysis, phosphorylation, and acetylation.[24] These proteins may be involved in different biological functions.[25,26] Breast cancer is a well-studied carcinoma through salivary proteomics providing a number of possible biomarkers by SELDI-TOF-MS used for initial detection and/or follow- up screening.[27] Overexpression of epidermal growth factor (EGF), cancer antigen 15-3 (CA15-3), and c-erbB-2 was significantly observed in breast cancer patient’s saliva as compared to healthy controls.[28-30] Salivary fluid protein vascular endothelial growth factor (VEGF), EGF, and carcinoembryonic antigen levels were found significantly overexpressed in saliva of breast cancer patients with high sensitivity and specificity.[31] Proteomic and transcriptomic signatures carbonic anhydrase VI (CA6) and psoriasin from saliva for the non-invasive detection of breast cancer may serve as biomarkers with high specificity and sensitivity using Affymetrix HG-U133-Plus-2.0 Array and two-dimensional difference gel electrophoresis (2D-DIGE).[32] Salivary proteins having phenylalanine and tryptophan have great potential for label-free and non-invasive detection of breast cancer by surface-enhanced Raman spectroscopy (SERS).[33] In lung cancer patients, upregulation of salivary proteins, zinc α2-glycoprotein (AZGP1), haptoglobin hp2 (HP), and human calprotectin (CALPR) were reported with high sensitivity and specificity through 2D-DIGE followed by MALDITOF as compared to healthy controls.[34] Therefore, salivary biomarker seems very promising for early detection and pathogenesis of disease[35] and use of saliva as an investigative tool is of great interest.
SIGNIFICANCE OF SCREENING BIOMARKERS IN OVARIAN CANCER
Asymptomatic nature at an initial stage and heterogeneity makes ovarian cancer a “silent killer” which is being considered as the most lethal gynecological malignancy by acquiring fifth foremost cause of cancer-related deaths in women. Despite having different subtypes with a collection of neoplasms with diverse clinic-pathological and molecular topographies, ovarian cancer is treated as a single disease.[36] Globally, ovarian cancer reached at a new height by attending seventh most common cancer with 240,000 new cases in 2018[37] and the second most common cancer after breast cancer over the age of 40 years in developed countries.[38]
It is unfortunate that only 20% of patients with ovarian cancer are getting diagnosed at their initial stage and majority of patients enter into advanced or metastatic stage due to lack of significant symptoms related to tumorigenesis and robust biomarkers.[39-41] Multiple molecular and proteomic signatures either from body fluid or tissues are being used for the detection and prognosis of ovarian cancer. However, majorly relies on screening with serum biomarkers such as gold standard CA125, human epididymis protein-4 (HE4), CA15-3, CA19-9, CA72-4, and macrophage colony-stimulating factor[42,43] and/or transvaginal ultrasound-imaging [Figure 3].[44] Advanced diagnostic techniques for the detection of ovarian cancer through, multiple blood tests and biopsies along with computed tomography (CT) and magnetic resonance imaging are costly, their radiation exposure restrict the use for screening purpose and invasive procedures also create an annoying condition for patients.[45]
- Multiple molecular and proteomic signatures either from body fluid or tissues are being used for the detection and prognosis of ovarian cancer.
Stage of ovarian cancer acts as powerful prognostic variable for expecting outcome after diagnosis. Initial diagnosis of the disease has a significant impact on mortality by helping to manage the disease with minimum discomfort of patient. Ovarian cancer can be cured up to 90%, if diagnosed early (Stage I) with confined malignancy to the ovaries only. If malignancy is restricted to the pelvis (Stage II), then also 5-year survival rate is 70% and 47.4% ovarian cancer patients can survive even after 5 years.[38] Survival rate decreases with the progression of disease and 5 years survival rate of advance stage patient (Stage III-IV) is only 20% or even less than that. Early diagnosis of disease will also help to improve the survival rate of patient by 10–30%[46,47] behaving as a hallmark of successful treatment. Detection of ovarian cancer through non-invasive procedure at initial stage will reduce patient discomfort by helping in prognosis and their therapeutic intervention. Therefore, a non-invasive and cost-effective procedure for the screening of ovarian cancer is much awaited.
BIOMARKERS OF OVARIAN CANCER (EXCEPT SALIVA)
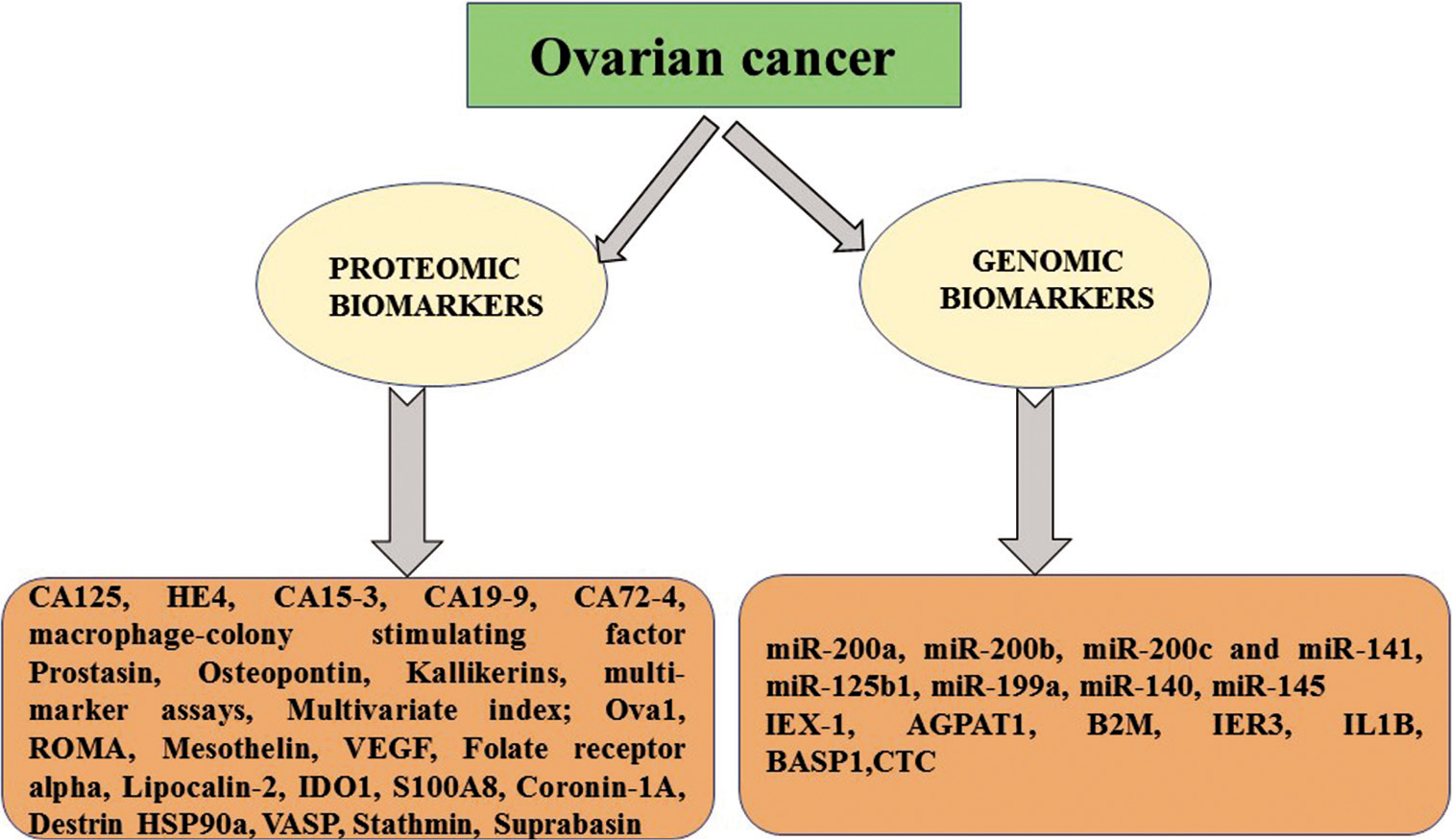
Biomarkers are much appreciated and effective tool for the detection of a disease at initial stage. The molecules produced either from tumor itself or its surrounding cells which can be measured either from body fluids and/or tissues for screening purpose are called tumor biomarkers. They are distinct and independent entities within human body and provide important information regarding the current status of physiologic condition of the body. Any evaluated indicator either from blood, urine, saliva, or tissues will be useful in determining the probability or stages of disease by providing a sign of normal or abnormal status of health. Evaluation of promising biomarkers from biological fluid or tissues creates new prospects in ovarian cancer detection and therapy by helping in risk assessment, diagnosis, and prognosis by quantitative measurement.[48] Identification of ovarian cancer at earliest stage is the epic success of the treatment in terms of survival rate that can be improved with better clinical outcomes. Serum CA-125 levels are regarded as the gold standard and widely used as a preoperative biomarker for ovarian cancer screening[49] whose overexpression correlates with poor survival rate.[50] Although, overexpression of CA125 is also found in some benign and non-gynecological conditions which refer to its non-specificity and less sensitivity as a reliable biomarker for screening of ovarian cancer.[51] Since only 80% of ovarian cancers and 50% of early stage patients express upregulation of CA125, multiple biomarkers are needed to enhance specificity and sensitivity for the detection of disease at early stage. Due to the biomarker’s inadequacy, it is required to combine CA125 with other markers/imaging to improve performance for the screening purpose. Researchers are now developing biomarkers complimentary to CA125 to aid in and use like multimarker assays. Several potential serum biomarkers such as HE4 and kallikreins have been evaluated alone and in combination with CA125, surprisingly with the combination of CA125, sensitivity and specificity of these proteins were increased.[52] Protein biomarker HE4 is approved by FDA as a tumor marker for the onset, progression, and reoccurrence of epithelial ovarian cancer.[53] HE4 either gives higher specificity 95% and sensitivity 73% for ovarian cancer screening, whereas in combination with CA125, sensitivity and specificity increase up to 100% and 74%, respectively.[54] Similarly, prostasin, osteopontin, and kallikreins outperform for screening of ovarian cancer when used alone, but their sensitivity and specificity increase remarkably in combination with CA125. Protein biomarkers mesothelin VEGF and folate receptor alpha (FOLR1)[50] also proved themselves a promising biomarker for ovarian cancer which will help to improve the clinical outcomes or long-term survival of patients.
A multivariate index assay (MIA) of 5-protein biomarker panel Ova1 is an algorithm to differentiate between cancerous and non-cancerous pelvic masses[55,56] in combination with second-generation CA125-II and other transport and inflammatory proteins such as transthyretin, apolipoproteinA-1,β2-Microglobulin, and transferrin. Highly sensitive (94%) Ova1 gives positive predictive value (PPV) of 40%, whereas negative predictive value is 93% for ovarian cancer.[57] A serum-based scoring system risk of ovarian malignancy algorithm is used for the assessment of low or high risk of a malignancy of pelvic mass for menopausal status as a combination of CA-125 and HE4. It gives 74.9% specificity and 93.8% sensitivity in postmenopausal women, whereas, in premenopausal condition, sensitivity and specificity were achieved 100% and 74.2%, respectively. Therefore, MIA seems a better option for pre-operative testing as compared to a single biomarker due to increased sensitivity.[58]
Tumorigenesis is a multistep process in which genetic alteration takes place in oncogenes as mutation, deletion or amplification, and changes in miRNA genes. miRNAs are considered as positive predictive biomarkers for screening and monitoring of different types of malignancy including ovarian cancer due to outstanding stability and resistance to degradation ability.[59-63] They are most profuse classes of gene regulatory molecules that circulate uninterruptedly in our body after secretion from specific cells to the recipient cells.[64-66] Expression of several genes that involved in various cellular processes associated with systemic diseases regulated by miRNAs[67,68] which are highly involved in the initiation and metastatic stage of carcinoma.[69,70] Any altered expression is closely related to the incursion and progression of disease. During the progression of disease, miRNA behaves in dual manner either as oncomiRs or tumor suppressive depending on their expression pattern and functions.[71] Aberrant expression of miRNAs either as upregulated or downregulated profile has been observed in various human malignancies such as lymphoma breast cancer, colorectal cancer, prostate cancer, and glioma.[72-76] Differential expression of miRNAs is also obvious in ovarian cancer either as cancer-promoting genes or as tumor suppressor genes, respectively, significant overexpression of miR-200a, miR-200b, miR-200c, and miR-141 and underexpression of miR-125b1, miR-199a, miR-140, and miR-145 were observed in ovarian cancer tissue as compare to normal tissue.[77] Downregulated expression of miR-31 in serous ovarian tumors and cell lines signifying as a tumor suppressor gene in ovarian cancer.[78] Altered expression of miRNAs can directly reflect the genomic or chromosomal changes such as genomic mutations, chromosomal abnormalities, epigenetic changes, and irregularities in miRNA biogenesis of cancer-associated genes.[79] These genomic changes can be uncover by sequencing technology which will help to identify genetic biomarkers for screening, staging, prognosis, and diagnosis of disease. Genetic biomarkers from saliva such as circulating tumor cells and circulating DNA/RNA will help to detect the disease non-invasively at early stage.[49] Regardless of several number of current and conventional methods which are used for the identification of ovarian cancer through multiple invasive procedure, it is not getting detected at earliest stage and survival of the patient is not being hassle free due to poor management of disease. The identification and validation of highly specific biomarker for ovarian cancer at initial stage are needed to establish by non-invasive screening methods.
BIOMARKERS OF OVARIAN CANCER IN SALIVA
Genomic biomarkers
Non-invasive techniques for screening and diagnosis of a disease by circulating tumor cells (CTCs), circulating tumor nucleic acids DNA/RNA, and exosomes are achieving a status of potential predictive and prognostic markers for malignancy. Gene transcripts present in each cell which discloses about the molecular and functional constituent, development and their quantity for a specific physiological condition are transcriptome.[80] Salivary transcriptomics are creating a new paradigm for non-invasive molecular diagnosis for systemic diseases through transcriptomic analysis using qPCR, microarray analysis, and sequencing techniques from saliva as micro-RNAs (miRNAs), circular RNAs, piwi-interacting RNAs, and non-coding RNAs.[81] Among all these salivary extracellular RNA, miRNAs are the excellent transcriptomic biomarkers due to their resistance to degradation ability and outstanding stability for early detection and monitoring of different types of malignancy including ovarian cancer.[59-63] miRNAs are nucleic acids encoded by genes which never get translated into proteins. Single-stranded, highly conserved, short segments of RNA molecule consisting of 21–25 nucleotides[82] found in all the 12 body fluids including saliva.[83] It is well documented that salivary miRNA behaves as a potential biomarker for breast cancer, lung cancer, and pancreatic cancer[32,84-86] with higher specificity and sensitivity. Salivary miRNAs consist of thousands of mRNAs and miRNAs[63] whose altered expression was observed in ovarian cancer patients by high-throughput technology microarray followed by RT-qPCR as a transcriptomic signature. Five salivary transcriptomic genes such as interleukin 1, beta, immediate early response 3 (IER3), beta-2-microglobulin, brain abundant, membrane attached signal protein 1, and 1-acylglycerol-3-phosphate O-acyltransferase 1 were significantly observed underexpressed in ovarian cancer patients with respect to healthy controls.[87] Early responsive gene X-1 (IEX-1) or IER3 is found to be involved in cell differentiation, growth, and apoptosis under cellular stress condition. Clinical potential of IEX-1 was validated in different groups of epithelial ovarian tumors and healthy control women using qRT-PCR in blood and saliva. A significant downregulation of IEX-1 in epithelial ovarian tumors was observed with high specificity and sensitivity that clearly indicate that IEX-1 behaves as a tumor suppressor gene in epithelial ovarian carcinoma.[88]
Due to increased apoptosis and necrosis in cancer cells, metabolic rate becomes very high and produces a large quantity of cell-free DNA (cfDNA) in blood, saliva, and urine.[89-91] These circulating cfDNAs help to detect genomic alteration associated with cancer.[92] Salivary DNA is highly stable and of good quality and can be used for the detection of circulating tumor-specific DNA (ctDNA) through “liquid biopsy”[93] whose differential expression corresponds initiation and progression of malignancies such as colon, breast stomach, and lung cancers.[5,94,95] Prognostic value of CTCs and ctDNA from saliva is well documented among breast, lung, colorectal, pancreatic, and gastric cancers patients in previous meta-analyses.[96] Shorter half-life of ctDNA helps to analyze tumor development, metastatic progression, and treatment efficacy in ovarian cancer.[92] However, prognostic value of CTCs was not associated with disease progression in ovarian cancer but found elevated with CA-125 and gave significant status of overall survival (OS), progression-free survival, and disease-free survival.[97] Therefore, RNA signatures in saliva may be a potential biomarker for ovarian cancer screening with significant sensitivity and specificity which may impact on current diagnostic procedures of ovarian cancer.
Proteomic biomarkers
Human protein atlas reports differential expression of salivary gland proteins from ductal epithelial cells, serous, or mucinous cells. Almost 60% (11,716) of all human proteins (19,613) are found differentially expressed, out of these total 85 salivary proteins were found overexpressed with respect to other tissue, where it is already reported. Salivary protein cystatin-SA (CST2), carbonic anhydrase VI (CA6), prolactin-induced protein, and amylase alpha 1B were over expressed in serous salivary glands, whereas mucin-7 (MUC7) is found upregulated mucinous salivary gland and sodium/iodide cotransporter (SLC5A5) in salivary ducts.[98] Differential expression of salivary proteins is analyzed by proteomic analysis using mass spectrophotometry that may serve as biomarkers to monitor onset and progression of ovarian cancer. The gold standard serum biomarker CA125 often called mucin 16 (MUC16) is single most biomarker approved by the US FDA for early diagnosis of ovarian cancer and disease monitoring.[58] This highly recommended serum biomarker is also found in saliva and gives a better diagnostic value with high specificity, diagnostic efficiency, and PPV than serum CA125 assay in ovarian cancer patients.[99,100] Therefore, trend of evaluating of CA125 levels from saliva through non-invasive approach for the screening of ovarian cancer would be a great initiative in this direction. Recently, differential expression of another salivary proteins such as indoleamine-2,3-dioxygenase1 (IDO1), lipocalin-2, and S100A8 was observed by fluorescence-based 2D-DIGE coupled with MALDI/ TOF-MS in ovarian cancer patient’s saliva as compare to healthy controls. Diagnostic potential of these biomarkers was evaluated by ROC analysis after further validation by Western blotting and ELISA, immunohistochemistry, and qRT-PCR in independent cohort. Specificity and sensitivity of these targeted salivary proteins were significantly high and have potential to be used as screening biomarker after further validation on large scale.[101]
Response of neoadjuvant drugs in ovarian cancer and organotropic behavior of breast and ovarian cancers was observed in saliva of metastatic ovarian and breast cancer patients. Differential expression of salivary proteins was analyzed by mass spectrometry in saliva of ovarian cancer patients getting chemotherapy. A well-documented protein HSP90a involved in cell proliferation and migration coronin-1A, and destrin was observed upregulated in saliva of ovarian cancer, and overexpression of proteins coronin-1A, VASP, suprabasin, and stathmin was also reported in ovarian cancer patients getting chemotherapy.[102] These metastatic-related proteins give an idea about their association with organotropism behavior of metastatic breast and ovarian cancers patients with/without chemotherapy.
Metabolomic biomarkers
Metabolomics is the study of chemical processes involving small molecule metabolites, intermediates, and end products of metabolism. Endogenous metabolites present in cell-free saliva have potential to act as biomarkers for onset and progression of various cancer types. Altered expression of human salivary metabolome was observed in physiological, environmental, and diseased conditions and potential salivary metabolomics markers were identified and validated for oral, breast, lung, pancreatic cancers, and periodontitis through various metabolic approaches.[5,102-104]
Oncogenesis is a multifaceted disease due to the variations in cellular pathways and several genes are involved in transforming normal cells into malignant forms. Biological pathways regulated by proteins provide energy and metabolites for cell differentiation and proliferation during tumorigenesis.[105] Proteins with covalently attached carbohydrate moieties such as hexose, fucose, hexosamine, and sialic acids (SAs) are called glycoproteins, which help to maintain healthy cellular life. However, overexpression of glycoproteins has been noticed during several pathological conditions.[106] Altered expression of protein glycosylation plays an important role in cancer progression and metastasis. In the saliva of breast cancer patients, some metabolites such as SA, proline, taurine, and valine were found overexpressed and considered potential biomarkers.[107] Upregulation of salivary metabolites including choline, pipecolinic acid, l-phenylalanine, and S-carboxymethyl-L-cysteine was reported in head-and-neck cancer, with excellent diagnostic test accuracy.[108] Furthermore, variation in expression levels of carbohydrate moieties of glycoproteins reflected the stage of cancer and might act as diagnostic tool for ovarian carcinoma.[109] Recently, it was reported that levels of SA in saliva make a significant differentiation between benign and cancer patients with higher specificity and sensitivity of 100% and 80%, respectively, through SERS. Therefore, SA can serve as a more reliable biomarker than CA-125 to detect ovarian cancer through non-invasive technique with a cutoff value of 15.5 mg/dL.[110] SA is a key monosaccharide present in body fluids, which constitute cell surface glycans on mammalian cells responsible for binding with glycolipids and glycoproteins and acts as important systematic biomarker of systemic inflammation.[111,112] Any alteration in terminal SA structures and changes in glycosylation positions are classic hallmarks of malignant transformation.[113] Abnormal protein glycosylation specifies conversion of normal cells in to the malignant forms by indicating high levels of serum glycoproteins, protein-bound hexoses, SA, hexosamine, and fucose in ovarian cancer patients in comparison to healthy controls.[114] SA, both alone or with hydroxyproline, in serum delivers a better diagnosis value than CA-125 and HE4 and can be used as a clinical interpreter for ovarian cancer. It was also observed that protein-bound SA has highest capacity to distinguish between ovarian cancer patients and healthy controls as compared to other glycoproteins present in serum.[114] Salivary metabolomics is currently in its nascent stage, especially in case of ovarian cancer as only SA is reported. To discover new and effective prognostic markers for diagnosis of ovarian cancer, salivary metabolomics can bring new possibilities.
FUTURE PROSPECTS
One of the deadliest cancers among women is ovarian cancer due to late diagnosis worldwide. Inter- and intra-heterogeneity makes difficult to manage ovarian cancer clinically. Multiple serial blood sampling and biopsies for monitoring the progression of the diseases are highly invasive for cancer patients. Therefore, a non-invasive technique for ovarian cancer screening and monitoring is prime necessity. Saliva is an obvious pool of tumor markers and salivaomics helps to understand the physiognomies of saliva with significant advancement on how the salivary constituents narrate their behavior and functions in pathogenesis. Although, salivaomics is in nascent stage, it is helping to identify various biomarkers for multiple malignancies. Massive efforts are also being done in both fundamental and clinical research to identify possible biomarkers from saliva that can reduce patient discomfort with positive outcome for ovarian cancer. Swiftly emerging technologies such as liquid biopsy, proteomics, and RNA sequencing have prospective to deliver innovative diagnostic solutions in the salivary field. These advanced technologies have broadly expanded the salivary diagnostic approach from the oral cancer to systemic diseases making salivary diagnostics a clinical reality. Precise and reliable biomolecules identified through salivaomics showing significant differences between malignant and normal healthy control may have potential to be used as a targeted drug or personalized medication for cancer patients.
Although, identification of a highly sensitive and specific biomarkers is major a challenge in saliva research due to variations in sample collection protocol, different quantitation methods, sample degradation, extensive range of molecular and cellular constituents, and lower protein concentrations. Discovery, verification, and validation phases of salivary biomarkers need validation in a larger sample size of independent cohort. Comparison of salivary biomarkers with other biological fluids or tissues will be important for determining its accuracy and feasibility to assess health and disease status. Effective use of targeted acquisition mass spectrometry strategies, through; selected reaction monitoring (SRM), multiple reaction monitoring, and parallel reaction monitoring, will help to achieve an accurate and reproducible quantification. SRM platforms should be executed in majority of biological and clinical laboratories to transform them into convenient and accurate POC diagnostics that can serve as a non-invasive diagnostic tool. With the help of SRM and POC, “lab-on-a-chip” approaches with constant improvement in NGS methods, liquid biopsy, advanced PCR-, and electromagnetic field-based technologies can be enabled for the diagnosis and monitoring of human diseases. These improvements in salivaomics will definitely help in early diagnosis of ovarian cancer patients and optimization of their treatment in future.
CONCLUSION
Saliva is widely accepted as an alternative source for biomarker discovery due to ease of sample collection and handling process. Identification of novel and robust salivary biomarkers via omics method can be used for early detection of ovarian cancer. Along with asymptomatic nature of ovarian cancer, social stigma also play important role for the late diagnosis of disease in large population. Diagnosis by saliva based tests will circumbent the social stigma as well as remove inhibition felt by women in getting tested with currently available tests. Therefore, saliva-omics has a possibility of becoming a very important tool for early detection of ovarian cancer.
Declaration of patient consent
Patient’s consent not required as patients identity is not disclosed or compromised.
Financial support and sponsorship
Authors acknowledge Indian Council of Medical Research, India, for the financial support (No.5/13/16/2013/NCD-III).
Conflicts of interest
There are no conflicts of interest.
References
- Review: The physiology of saliva and transfer of drugs into saliva. Forensic Sci Int. 2005;150:119-31.
- [Google Scholar]
- Salivary secretion: Mechanism and neural regulation. Monogr Oral Sci. 2014;24:14-29.
- [Google Scholar]
- Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin Microbiol Rev. 2013;26:781-91.
- [Google Scholar]
- Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med. 2012;4:82.
- [Google Scholar]
- A human scFv antibody generation pipeline for proteome research. J Biotechnol. 2011;152:159-70.
- [Google Scholar]
- Application of saliva for drug monitoring An in vivo model for transmembrane transport. Eur J Clin Chem Clin Biochem. 1996;34:171-91.
- [Google Scholar]
- Therapeutic drug monitoring in saliva. An update. Clin Pharmacokinet. 1992;23:365-79.
- [Google Scholar]
- Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol. 2018;10:1-24.
- [Google Scholar]
- Potential applications of human saliva as diagnostic fluid. Acta Otorhinolaryngol Ital. 2011;31:347-57.
- [Google Scholar]
- Salivary biomarkers for the diagnosis and monitoring of neurological diseases. Biomed J. 2018;41:63-87.
- [Google Scholar]
- Saliva diagnostics: Utilizing oral fluids to determine health status. Monogr Oral Sci. 2014;24:88-98.
- [Google Scholar]
- Network generation enhances interpretation of proteomics data sets by a combination of two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ ionization time of flight mass spectrometry. Analyst. 2012;137:4703-11.
- [Google Scholar]
- Proteome and proteomics: New technologies, new concepts, and new words. Electrophoresis. 1998;19:1853-61.
- [Google Scholar]
- Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl. 2009;3:116-34.
- [Google Scholar]
- Proteomic analysis of saliva: 2D gel electrophoresis, LC-MS/MS, and western blotting. Methods Mol Biol. 2010;666:31-41.
- [Google Scholar]
- Mass spectrometry-based clinical proteomics: Head-and-neck cancer biomarkers and drug-targets discovery. Mass Spectrom Rev. 2010;29:945-61.
- [Google Scholar]
- Saliva analysis by surface-enhanced laser desorption/ ionization time-of-flight mass spectrometry (SELDI-TOF/MS): From sample collection to data analysis. Clin Chem Lab Med. 2008;46:89-99.
- [Google Scholar]
- Protein posttranslational modifications: The chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342-72.
- [Google Scholar]
- Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome. Genome Med. 2016;8:44.
- [Google Scholar]
- Quantitative salivary proteomic differences in oral chronic graft-versus-host disease. J Clin Immunol. 2012;32:1390-9.
- [Google Scholar]
- The use of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to detect putative breast cancer markers in saliva: A feasibility study. J Oral Pathol Med. 2006;35:292-300.
- [Google Scholar]
- Epidermal growth factor in plasma and saliva of patients with active breast cancer and breast cancer patients in follow-up compared with healthy women. Breast Cancer Res Treat. 1997;42:83-6.
- [Google Scholar]
- The presence of soluble c-erbB-2 in saliva and serum among women with breast carcinoma: A preliminary study. Clin Cancer Res. 2000;6:2363-70.
- [Google Scholar]
- A preliminary study of CA15-3, cerbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Invest. 2000;18:101-9.
- [Google Scholar]
- Salivary protein factors are elevated in breast cancer patients. Mol Med Rep. 2008;1:375-8.
- [Google Scholar]
- Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS One. 2010;5:e15573.
- [Google Scholar]
- Surface-enhanced Raman spectroscopy of saliva proteins for the non-invasive differentiation of benign and malignant breast tumors. Int J Nanomedicine. 2015;10:537-47.
- [Google Scholar]
- Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol Cell Proteomics. 2012;11:1-12.
- [Google Scholar]
- Advances in ovarian cancer proteomics: The quest for biomarkers and improved therapeutic interventions. Expert Rev Proteomics. 2008;5:551-60.
- [Google Scholar]
- Current state of biomarkers in ovarian cancer prognosis. Future Oncol. 2015;11:3187-95.
- [Google Scholar]
- National Cancer Institute Surveillance, Epidemiology, and End Results Program Cancer Stat facts Ovarian Cancer. Maryland: National Cancer Institute; 2018.
- [Google Scholar]
- Targeting genetic and epigenetic alterations in the treatment of serous ovarian cancer. Cancer Genet. 2011;204:525-35.
- [Google Scholar]
- OVX1, macrophage-colony stimulating factor, and CA-125-II as tumor markers for epithelial ovarian carcinoma: A critical appraisal. Cancer. 2001;92:2837-44.
- [Google Scholar]
- Combined use of biomarkers for detection of ovarian cancer in high-risk women. Tumour Biol. 2010;31:209-15.
- [Google Scholar]
- HE4 and CA125 as a diagnostic test in ovarian cancer: Prospective validation of the risk of ovarian malignancy algorithm. Br J Cancer. 2011;104:863-70.
- [Google Scholar]
- Clinical implications and future perspectives of circulating tumor cells and biomarkers in clinical outcomes of colorectal cancer. Transl Oncol. 2016;9:340-7.
- [Google Scholar]
- Quantifying the potential benefit of CA 125 screening for ovarian cancer. J Clin Epidemiol. 1991;44:365-80.
- [Google Scholar]
- Estimating cost-effectiveness of a multimodal ovarian cancer screening program in the United States: Secondary analysis of the UK collaborative trial of ovarian cancer screening (UKCTOCS) JAMA Oncol. 2018;4:190-5.
- [Google Scholar]
- Applying genomics to organ transplantation medicine in both discovery and validation of biomarkers. Int Immunopharmacol. 2007;7:1948-60.
- [Google Scholar]
- Prognostic importance of preoperative CA-125 in international federation of gynecology and obstetrics stage I epithelial ovarian cancer: An Australian multicenter study. J Clin Oncol. 2005;23:5938-42.
- [Google Scholar]
- The lack of significance of Ca125 response in epithelial ovarian cancer patients treated with neoadjuvant chemotherapy and delayed primary surgical debulking. Gynecol Oncol. 2007;105:712-5.
- [Google Scholar]
- A perspective on ovarian cancer biomarkers: Past, present and yet-to-come. Diagnostics (Basel). 2017;7:14-22.
- [Google Scholar]
- HE4 in ovarian cancer: From discovery to clinical application. Adv Clin Chem. 2011;55:1-20.
- [Google Scholar]
- Evaluation of the diagnostic accuracy of the risk of ovarian malignancy algorithm in women with a pelvic mass. Obstet Gynecol. 2011;118:280-8.
- [Google Scholar]
- Effectiveness of a multivariate index assay in the preoperative assessment of ovarian tumors. Obstet Gynecol. 2011;117:1289-97.
- [Google Scholar]
- The road from discovery to clinical diagnostics: Lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev. 2010;19:2995-9.
- [Google Scholar]
- Performance of the American college of obstetricians and gynecologists’ ovarian tumor referral guidelines with a multivariate index assay. Obstet Gynecol. 2011;117:1298-306.
- [Google Scholar]
- Emerging diagnostic, prognostic and therapeutic biomarkers for ovarian cancer. Cell Oncol (Dordr). 2017;40:105-18.
- [Google Scholar]
- MicroRNA history: Discovery, recent applications, and next frontiers. Mutat Res. 2011;717:1-8.
- [Google Scholar]
- Role of microRNAs as Clinical cancer biomarkers for ovarian cancer: A short overview. Cells. 2020;9:169-89.
- [Google Scholar]
- Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS One. 2013;8:e57502.
- [Google Scholar]
- Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev Res (Phila). 2015;8:165-73.
- [Google Scholar]
- Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34-38.
- [Google Scholar]
- Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442-52.
- [Google Scholar]
- Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-9.
- [Google Scholar]
- MicroRNAs in the search for understanding human diseases. BioDrugs. 2007;21:97-104.
- [Google Scholar]
- MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget. 2016;7:6476-505.
- [Google Scholar]
- Circulating microRNAs as biomarkers of colorectal cancer: Results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol. 2013;11:681-8.e3.
- [Google Scholar]
- MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006;66:7390-4.
- [Google Scholar]
- Role of microRNA deregulation in the pathogenesis of diffuse large B-cell lymphoma (DLBCL) Leuk Res. 2013;37:1420-8.
- [Google Scholar]
- MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One. 2014;9:e96228.
- [Google Scholar]
- MicroRNA dysregulation in colorectal cancer: A clinical perspective. Br J Cancer. 2011;104:893-8.
- [Google Scholar]
- MicroRNAs and prostate cancer: From preclinical research to translational oncology. Cancer J. 2012;18:253-61.
- [Google Scholar]
- Circulating microRNA biomarkers for glioma and predicting response to therapy. Mol Neurobiol. 2014;50:545-58.
- [Google Scholar]
- Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906-15.
- [Google Scholar]
- MicroRNAs as potential biomarkers in cancer: Opportunities and challenges. Biomed Res Int. 2015;2015:125094.
- [Google Scholar]
- RNA sequencing analysis of salivary extracellular RNA. Methods Mol Biol. 2017;1537:17-36.
- [Google Scholar]
- MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522-31.
- [Google Scholar]
- Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci. 2012;69:3341-50.
- [Google Scholar]
- Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology. 2010;138:949-57.
- [Google Scholar]
- MicroRNA expression in salivary supernatant of patients with pancreatic cancer and its relationship with ZHENG. Biomed Res Int. 2014;2014:756347.
- [Google Scholar]
- Salivary transcriptomic biomarkers for detection of ovarian cancer: For serous papillary adenocarcinoma. J Mol Med (Berl). 2012;90:427-34.
- [Google Scholar]
- Significance of blood and salivary IEX-1 expression in diagnosis of epithelial ovarian carcinoma. J Obstet Gynaecol Res. 2018;44:764-71.
- [Google Scholar]
- DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659-65.
- [Google Scholar]
- The translational potential of circulating tumour DNA in oncology. Clin Biochem. 2015;48:957-61.
- [Google Scholar]
- Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med. 2014;190:1117-26.
- [Google Scholar]
- Quantification of somatic chromosomal rearrangements in circulating cell-free DNA from ovarian cancers. Sci Rep. 2016;6:29831.
- [Google Scholar]
- Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One. 2015;10:e0145754.
- [Google Scholar]
- Quality and quantity of saliva DNA obtained from the self-administrated oragene method-a pilot study on the cohort of Swedish men. Cancer Epidemiol Biomarkers Prev. 2006;15:1742-5.
- [Google Scholar]
- Quantity and quality assessment of DNA extracted from saliva and blood. Clin Lab. 2012;58:307-12.
- [Google Scholar]
- Prognostic value of circulating tumor cells in ovarian cancer: A meta-analysis. PLoS One. 2015;10:e0130873.
- [Google Scholar]
- The Role of Circulating Biomarkers in the Early Diagnosis of Ovarian Cancer, Ovarian Cancer-From Pathogenesis to Treatment, Omer Devaja and Andreas Papadopoulos London: InTech Open; 2018.
- [Google Scholar]
- Saliva and serum CA 125 assays for detecting malignant ovarian tumors. Obstet Gynecol. 1990;75:701-4.
- [Google Scholar]
- Relationship between saliva and serum CA 125 in women with and without epithelial ovarian cancer. Obstet Gynecol. 1993;81:989-92.
- [Google Scholar]
- Identification and validation of salivary proteomic signatures for non-invasive detection of ovarian cancer. Int J Biol Macromol. 2018;108:503-14.
- [Google Scholar]
- In search of the altering salivary proteome in metastatic breast and ovarian cancers. FASEB Bioadv. 2019;1:191-207.
- [Google Scholar]
- Branched-chain amino acid metabolism in cancer. Curr Opin Clin Nutr Metab Care. 2018;21:64-70.
- [Google Scholar]
- Increasing the sialylation of therapeutic glycoproteins: The potential of the sialic acid biosynthetic pathway. J Pharm Sci. 2009;98:3499-508.
- [Google Scholar]
- Salivary biomarkers in the diagnosis of breast cancer: A review. Crit Rev Oncol Hematol. 2017;110:62-73.
- [Google Scholar]
- Diagnostic capability of salivary biomarkers in the assessment of head and neck cancer: A systematic review and meta-analysis. Oral Oncol. 2015;51:805-18.
- [Google Scholar]
- A case control study of glycoprotein status in ovarian carcinoma. Clin Biochem. 2005;38:535-9.
- [Google Scholar]
- Determination of sialic acid in saliva by means of surface-enhanced Raman spectroscopy as a marker in adnexal mass patients: Ovarian cancer vs benign cases. J Ovarian Res. 2018;11:61.
- [Google Scholar]
- Rosenberg A In: Biology of the Sialic Acids. New York: Plenum Press; 1995. p. :7-49.
- [Google Scholar]
- Serum levels of glycoproteins are elevated in patients with ovarian cancer. Indian J Clin Biochem. 2014;29:345-50.
- [Google Scholar]
- Combined detection of sialic acid and hydroxyproline in diagnosis of ovarian cancer and its comparison with human epididymis protein 4 and carbohydrate antigen 125. Clin Chim Acta. 2015;439:148-53.
- [Google Scholar]






