Translate this page into:
Identification of testis derived differentially expressed spermatogenic microRNAs in idiopathic hypospermatogenesis

*Corresponding author: Manish Jain, Department of Reproductive Biology, All India Institute of Medical Sciences, New Delhi, India. manishjain@aiims.edu
-
Received: ,
Accepted: ,
How to cite this article: Sharma N, Halder A, Kaushal S, Kumar M, Jain M. Identification of testis derived differentially expressed spermatogenic microRNAs in idiopathic hypospermatogenesis. J Reprod Health Med. 2025;6:4. doi: 10.25259/JRHM_30_2024
Abstract
Objectives
Hypospermatogenesis (HS) is a common histopathological subtype of non-obstructive azoospermia (NOA), characterized by a decrease in the total number of germ cells within the seminiferous tubule. The diagnosis of HS is made by invasive procedures like testicular biopsy or fine needle aspiration (FNA). MicroRNAs (miRs) are biomolecules with emerging roles as diagnostic biomarkers for diseases. This study aimed to investigate the differential miR expression profile in idiopathic HS (iHS) to explore candidate spermatogenic miRs in human male infertility.
Material and Methods
In this observational study, patients reporting azoospermia due to spermatogenic failure were recruited prospectively. Genetic exclusion was performed using XY-fluorescent in-situ hybridization (XY-FISH) and Yq microdeletion. Testicular FNA samples were used for total ribonucleic ccid (RNA) isolation. Small RNA sequencing-based exploratory analysis was performed on 20 iHS patients and five normospermatogenesis (NS) patients. nCounter miRNA expression based validation was performed for four iHS and four NS patients.
Results
Analysis of miRs in testicular tissue showed differential expression patterns having 49 downregulated and 3 upregulated miRs between iHS and NS patients, with miR-379-5p, miR-449a, miR-181c, miR-34b-3p, and miR-122b-5p being notable candidate spermatogenic miRNAs. Pathways such as Phosphatidylinositol 3 Kinase-Protein Kinase B (PI3K-Akt) and mitogen-activated protein kinase (MAPK) signaling and molecular functions like apoptosis and cell differentiation were significantly enriched in iHS patients. This is the first study of its kind to investigate the differential expression of micro-ribonucleic acid (miRs) in a cohort of NOA patients exclusively of the HS subtype. Comparing the study results to previously published data revealed that dysregulated spermatogenic miRs are shared in HS and other NOA subtypes. The analysis of spermatogenic miRs according to each patient’s profile showed significant dysregulation in miR expression, linked to 40% of cases with idiopathic HS.
Conclusion
This study provides important insight into the potential of miRNA to be used as a biomarker for the diagnosis of iHS. Although the study is based on a relatively low sample size, it provides a proof of concept which can be validated in a larger cohort.
Keywords
Candidate spermatogenic microRNAs
Differential microRNAs expression
Hypospermatogenesis
INTRODUCTION
As infertility rates rise globally, there has been a growing recognition of the significant role of male factors in its pathogenesis.[1,2] Azoospermia is the condition in which no sperm is seen even after centrifugation of the semen samples. Spermatogenic failure giving rise to non-obstructive azoospermia (NOA) can be evaluated by histopathological examination of testes and discern three subtypes: hypospermatogenesis (HS), Sertoli cell only syndrome (SCOS), and maturation arrest (MA).[3] Approximately 45% of NOA patients exhibit identifiable causes, with the remainder being idiopathic.[4] HS is reportedly the second most prevalent subtype of NOA in which the overall number of germ cells at all the developmental stages is notably reduced.[5] The primary defect in HS lies within the stem cell population, making it a complex and multifactorial condition with the involvement of both genetic and epigenetic factors. Understanding the epigenetic basis of HS is essential not only for diagnosis but also for family planning, as there is a substantial risk of transmitting these factors to offspring. Despite its prevalence and clinical significance, the mechanisms underlying idiopathic HS (iHS) remain largely elusive, which poses significant diagnostic and therapeutic challenges. Several studies have identified dysregulated microRNAs (miRs) that may contribute to the pathogenesis of HS. Noveski et al. examined the differential expression (DE) profile of miRs in testicular biopsies from patients with spermatogenesis disorders compared to normal spermatogenesis. They identified five deregulated miRs, including hsa-miR-34b, hsa-miR-449b, hsa-miR-517c, hsa-miR-181c, and hsa-miR-605, which have involvement in pathways like tumor necrosis factor-related apoptosis-inducing ligand signaling.[5] Lian et al. studied microarray-based miR profiles of specifically early MA patients.[6] Zhang et al. have further studied the miR profiles and commented on the diagnostic specificity of upregulated (miR-370-3p, miR-539-5p, miR-10b-3p, and miR-22-5p) and four downregulated (miR-34b-5p, miR-31-5p, miR-516b-5p, and miR-122-5p) miRs.[7] Abu-Halima et al. studied SCOS and MA subtypes, comparing normospermatogenic samples for DE profiles of miRNAs. The SCOS and MA subtypes of NOA are most extensively studied using NGS techniques for miR profiling, whereas, data on the HS subtype are not available.[8] The presence of HS in NOA patients needs careful examination, as it may impact the prognosis for successful sperm retrieval and subsequent fertility treatments such as in vitro fertilization/intracytoplasmic sperm injection. Therefore, the present study aims to identify the DE profile of miRs associated with iHS. Testicular tissue-derived miR profiling done on NOA patients in previous studies has been compared with the findings of our study, and a panel of candidate spermatogenic miRs has been identified. The data generated from these analyses are used to understand the potential miR candidates of diagnostic importance and regulatory mechanisms associated with HS, which will ultimately contribute to the understanding of male infertility. This will facilitate the development of more effective diagnostic strategies and personalized treatment modalities for HS patients.
MATERIAL AND METHODS
Patient recruitment
The study was conducted at the Department of Reproductive Biology, All India Institute of Medical Sciences (AIIMS), New Delhi, India, for 3 years, from March 2020 to March 2023. A total of 50 infertile male patients coming to the department of urology were recruited. Clinical history collection and physical examination were performed for each case. Informed, written consent was obtained from the participants, and the study was performed according to the Declaration of Helsinki. Patients were excluded if they had conditions such as the presence of hydrocele or varicocele, history of testicular trauma, testicular maldescent, and history of radiotherapy, chemotherapy, or exposure to any reproductive toxin and orchitis.
Semen analysis
Semen analysis was performed for each infertile man following WHO criteria 2021. The ejaculate was collected in a pre-weighed container, and semen volume was calculated. A 30-min incubation at room temperature was followed for liquefaction of the semen sample, and pH was determined. The semen sample was heated in strong acid and resorcinol to measure fructose concentration. A 20 μL semen sample was prepared by carefully mixing the ejaculate and examined under ×10 followed by ×40 magnifications. For spermatozoa ≥101 per field at ×40 magnification, a 1:20 dilution ratio was chosen to count sperm in 50 μL of well-mixed ejaculate and 950 μL of fixative. Microscopic counting of spermatozoa was done in Neubauer’s chamber. Cases with no sperm seen even after centrifugation and a sperm count <2.5 million were considered azoospermia and oligozoospermia, respectively. The patients with obstructive causes were excluded, and a bilateral testicular fine-needle aspiration (FNA) evaluation was performed for a confirmatory diagnosis of NOA.
Hormonal evaluation
Hormonal evaluation of infertile men was performed using Chemiluminescent Microparticle Immunoassay. Quantification of serum follicle-stimulating hormone, serum luteinizing hormone, and serum testosterone was performed using 2nd generation Abbott laboratories’ kits.
Identification of HS cases
Azoospermic and oligozoospermic men with normal or slightly altered reproductive endocrine parameters were advised for testicular FNA evaluation to be categorized as HS. The testicular FNA procedure followed by reporting was done at the Department of Pathology. To obtain the sample, a spermatic block containing 1% lignocaine was given to the patients. A 22-gauge needle attached to a 10 mL syringe on a plunger was used at two distinct locations to get the aspirates from the right and left testicles. A thread-like structure in the aspirate was used to prepare smears before staining. Every smear was confirmed for the presence of at least 2000 cells. FNA from both testes showed a considerable decrease in the spermatogenic series of cells per Sertoli cell; however, maturation till spermatozoa was seen was considered to be suggestive of HS. The FNA findings showing the presence of normal spermatogenesis and mature spermatozoa were considered to be suggestive of normospermatogenesis (NS).
Identification of iHS cases
Blood samples of 50 HS patients [Supplementary Sheet 1] were collected in EDTA vials for DNA isolation using the QIAamp DNA Blood Mini Kit (QIAGEN, 51106) according to the manufacturer’s instructions. The optical density of the isolated DNA was measured with a spectrophotometer (Nanodrop 2100). Metaphase cell culture was conducted for 50 HS cases using a heparinized blood sample into karyotyping medium (Thermofisher Ref 12557-013). XY-fluorescent in situ hybridization (XY FISH) analysis was performed on metaphase cells utilizing in-house X and Y centromere probes. Signals from X- and Y-centromeric probes were enumerated in a minimum of five metaphase cells. Multiplex sequence tagged sites polymerase chain reaction (PCR) analysis for Yq microdeletion was carried out for all HS cases using AZFa- sY84, sY86; AZFb -sY127, sY134; AZFc -sY254, sY255, and sex-determining region Y (SRY) primers. The PCR amplicons were run on a 4% agarose gel and visualized. The HS cases were considered idiopathic after excluding microdeletions of the Y chromosome found in four patients and aneuploidies of the sex chromosomes by FISH analysis found in five patients. Total RNA isolation was performed for all HS and NS patients and RNA integrity number (RIN) calculation was found appropriate for 33 patients. We finally selected a total of 20 iHS cases and five controls for the differential miR expression using small RNA sequencing analysis, four iHS cases, and four NS controls for validation using nCounter miRNA expression analysis [Figure 1].
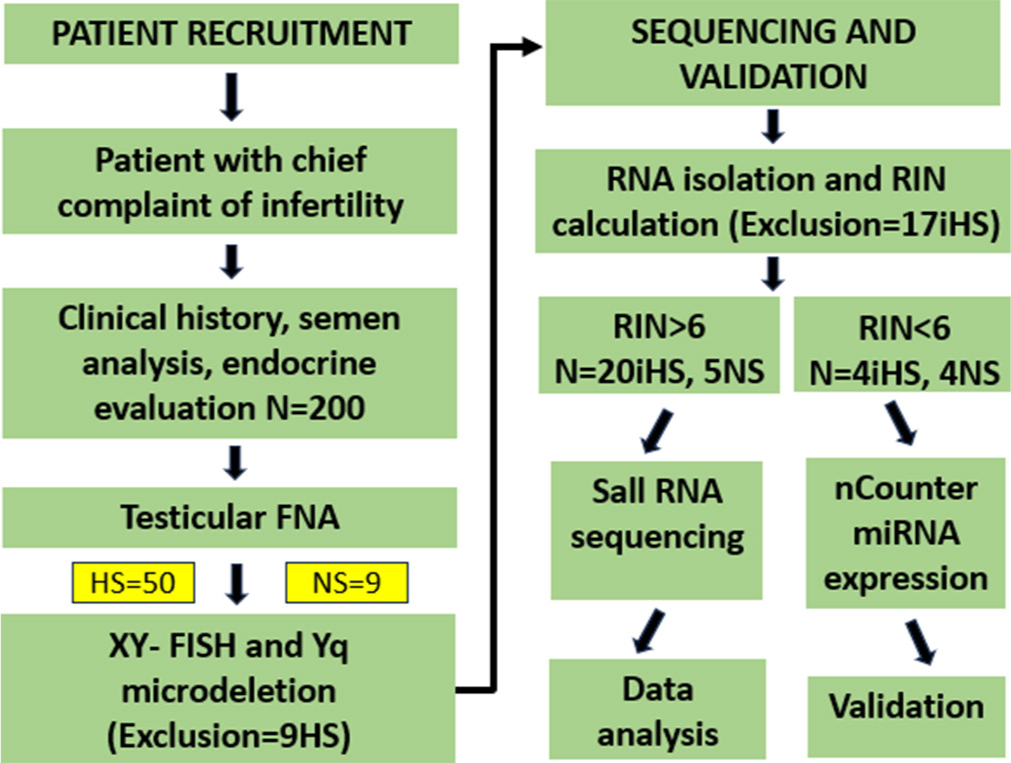
- Workflow of the study. RIN: Ribonucleic acid integrity number, RNA: Ribonucleic acid, XY-FISH: Fluorescence in-situ hybridization for X and Y chromosomes, HS: Hypospermatogenesis, iHS: Idiopathic hypospermatogenesis, NS: Normospermatogenesis
Small RNA sequencing and data analysis
Total RNA was extracted from testicular FNA samples of patients using the TRIzol method with a mirVana RNA isolation kit (Invitrogen, ThermoFisher Scientific-AM1560) according to the manufacturer’s protocol. The RNA concentration and integrity were assessed using the Qubit RNA Assay Kit on a Qubit 4.0 Fluorometer (Life Technologies) and the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system. We found 20 iHS cases with a RIN value of more than six and four cases with a RIN value of less than six. Further, the former was admissible for small RNA sequencing while later was used for validation using nCounter miRNA expression analysis. New England Biolabs (NEB) Next® Multiplex Small RNA Library Prep for Illumina was used to prepare the libraries. The 5’ adapter ligation was performed, followed by reverse transcription to synthesize first-strand complementary DNA. Prepared libraries were sequenced on Illumina NextSeq500 for 50 bp paired-end reads. Sequenced data were processed to generate fast quality (FASTQ) files. The adapter was trimmed with Cutadapt version 1.8dev. Bowtie2 was utilized for contamination removal (version 2.2.4). The reads were aligned to the human reference genome (version hg19) and mirBase version 21. Using mirDeep2, the known and novel miRNA predictions and their abundances were estimated. Differential estimation was performed by the edgeR package in RStudio. With log2fc > 1 and FDR 0.05, the up-and down-regulated miRNAs were calculated.
nCounter miRNA assay
The nCounter miRNA Expression Assay is designed to provide an ultra-sensitive, reproducible, and highly multiplexed method for detecting miRNAs in total RNA across all biological levels of expression. The assay was performed using the nCounter Analysis System (Nanostring Technologies). The assay provides a method for detecting miRNAs without the use of reverse transcription or amplification using molecular barcodes called nCounter Reporter Probes. It can potentially assay up to 800 different targets at once. nCounter miRNA assay is a 2 days procedure, with manual processing on day 1 and automated processing on day 2. Manual processing consists of miRNA sample preparation, miRNA Code Set Hybridization, and downstream processing. Automated processing consists of the Prep Station run or SPRINT run and Digital Analyzer for Data Collection (MAX/FLEX only). Four iHS and Four NS RNA samples that failed the Quality control test for miRNA sequencing, that is, RIN value less than six, were processed for nCounter miRNA assay.
In silico analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using DAVID. This tool calculates enrichment scores (e.g., Fisher’s exact test P-value and BenjaminiHochberg corrected P-value) to calculate the significance of enrichment for each GO term. An adjusted P < 0.05 was considered a statistically significant deviation from the expected distribution, and therefore, the corresponding GO term, as well as the KEGG pathway, was enriched in the target genes.
Ethical approval
The study was initiated after obtaining ethical approval (IEC-151/06.03.2020, RP-17/2020) from the Institute Ethical Committee (IEC) of AIIMS, India.
RESULTS
DE profile of miRs
The descriptive analysis of small RNA sequencing data was done by principal component analysis (PCA) of 20 iHS and five NS cases. In the PCA plot, it was observed that eight HS samples exhibited maximum variability in the data compared to five NS and 12 HS samples [Figure 2a]. However, the clustering of HS and NS samples did not occur in orthogonal coordinates, which suggests that there is no substantial difference in the expression pattern between the two groups. The volcano plot representation of significantly up- and down-regulated miRs is shown in Figure 2b, in which three and 49 miRs were found to be up-and down-regulated, respectively [Figure 2b]. The heatmap representation of up- and downregulated miRs is shown in Figure 2c. Individual data files for the DE miRs can be provided by the authors on request.
Identification of spermatogenic miRs
We performed a literature exploration to compare the current findings with existing DE miR profiles of NOA. We also searched for a panel of candidate miRs with strong evidence of causing spermatogenic failure in various subtypes such as SCOS and MA. The overlapping and non-overlapping miRs between NOA (SCOS and MA) and iHS groups were identified. Figure 2d shows the common miRs whose expression was altered in the literature-derived miRs profile of NOA and HS. Two miRs that were upregulated in NOA were found to be upregulated in HS. Similarly, 25 downregulated miRs in NOA were observed to be downregulated in HS. We found hsa-miR-379-5p as upregulated and hsa-miR-449a, hsa-miR-181c, hsa-miR-34b-3p, and hsa-miR-122b-5p as downregulated candidate spermatogenic miRs in 20 his patients. The distribution of all five miRs with respective log2 fold change values in eight out of 20 HS patients is shown in Table 1. The eight HS patients are shown to have significant difference in expression pattern when compared with controls and even the rest of the case, as depicted in Figure 2a. The clinical parameters of the patients are summarized in Table 2.
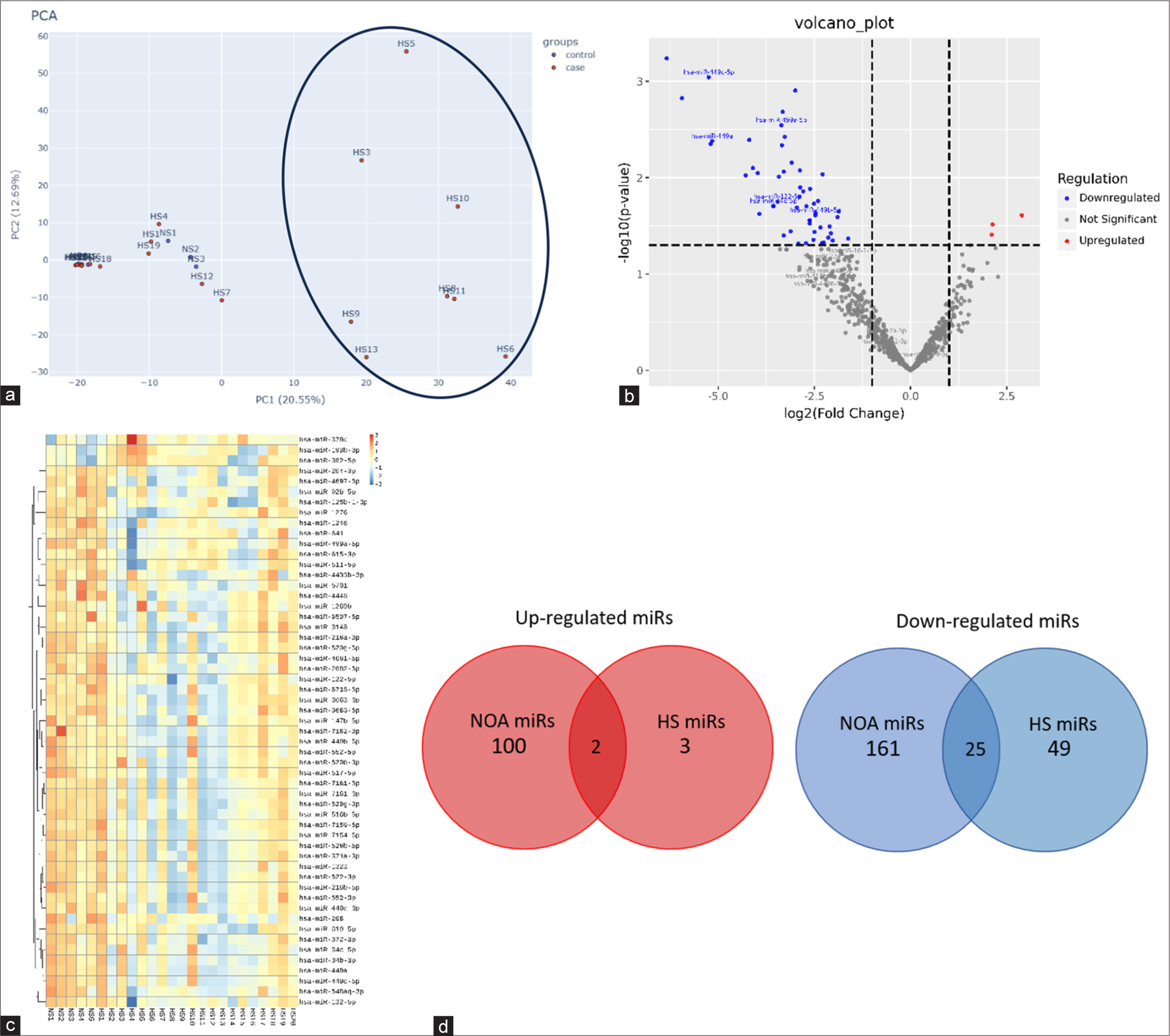
- Analysis of testicular miRs in iHS pathology: (a) Principal component analysis (PCA) plot showing 8 HS samples (marked with oval) showing maximum variability compared to the 5 NS samples, (b) Volcano plot of all significantly upregulated and downregulated miRs with Log2 Fold change cut-off >1 (c) Heatmap representation of DE miRs (49 downrwgulated and 3 upregulated) in 20 iHS and 5 NS patients. (d) Venn diagram showing common up and downregulated miRs in NOA versus iHS patients. PCA: Principle component analysis, NS: Normospermatogenesis, NOA: Non obstructive azoospermia, HS: idiopathic Hypospermatogenesis
| miRNAs | iHS3 | iHS5 | iHS6 | iHS8 | iHS9 | iHS10 | iHS11 | iHS13 |
|---|---|---|---|---|---|---|---|---|
| hsa-miR-379-5p | ↑2.14 | ↑2.0 | ||||||
| hsa-miR-449a | ↓−3.75 | ↓−3.85 | ↓−13.44 | ↓−2.53 | ||||
| hsa-miR-181c | ↓−9.82 | ↓−10.49 | ↓−2.14 | ↓−2.53 | ||||
| hsa-miR-34b-3p | ↓−13.94 | ↓−2.14 | ↓−3.59 | ↓−2.53 | ↓−3.44 | |||
| hsa-miR-122b-5p | ↓−4.9 | ↓−4.79 | ↓−6.7 | ↓−7.1 | ↓−5.5 |
Red up arrow: Upregulated miRs with fold change value, blue down arrow: Downregulated miRs with fold change value, differential estimation was done using the exactTest with the dispersion value <0.00001 through edgeR package in Rstudio, iHS: Idiopathic hypospermatogenesis, miR: MicroRNA
| Parameter | iHS3 | iHS5 | iHS6 | iHS8 | iHS9 | iHS10 | iHS11 | iHS13 |
|---|---|---|---|---|---|---|---|---|
| Age (year) | 34 | 30 | 27 | 37 | 28 | 24 | 27 | 35 |
| Duration of infertility | Primary infertility for 6 years | Primary infertility for 4 years | Primary infertility for 3 years | Primary infertility for 8 years | Primary infertility for 4 years | Primary infertility for 2 years | Primary infertility for 6 years | Primary infertility for 9 years |
| BMI | 25.5 | 27.4 | 24.2 | 26.7 | 25.3 | 23.4 | ||
| Semen analysis | Severe Oligo., Fructose +ve, Alkaline | Azoospermia, Fructose +ve, Alkaline | Azoospermia, Fructose +ve, Alkaline | Azoospermia, Fructose +ve, Alkaline | Azoospermia, Fructose +ve, Alkaline | Azoospermia, Fructose +ve, Alkaline | Azoospermia, Fructose +ve, Alkaline | Azoospermia, Fructose +ve, Alkaline |
| Serum T | 5 | 3.64 | 2.65 | 4.77 | 3.26 | 4.2 | 5.4 | 4.6 |
| Serum FSH | 7.59 | 19.69 | 28.95 | 3.7 | 6.8 | 10.48 | 12.14 | 16.43 |
| Serum LH | 4.37 | 5.35 | 6.63 | 6.08 | 3.45 | 2.67 | 4.45 | 3.7 |
| Testicular Vol. (mL) | R=15 L=15 |
R=12 L=12 |
R=12 L=12 |
R=15 L=15 |
R=15 L=12 |
R=12 L=10 |
R=14 L=12 |
R=15 L=14 |
| XY-FISH | N | N | N | N | N | N | N | N |
| Yq MD | N | N | N | N | N | N | N | N |
iHS: Idiopathic hypospermatogenesis, miR: MicroRNA, BMI: Body mass index, Serum T: Serum testosterone (reference range=1.42–9.23 ng/mL), Serum FSH: Serum follicle stimulating hormone (reference range=0.95–11.95 mIU/mL), Serum LH: Serum luteinizing hormone (reference range=0.57–12.07 mIU/mL), Testicular vol.=Testicular volume right and left, (Azoospermia=No sperm seen even after centrifugation, Severe oligo: Severe oligozoospermia [reference range <2.5 Million sperm in the ejaculate seen after centrifugation]); N: Normal phenotype, XY-FISH: XY Fluorescence in-situ hybridization, MD: Microdeletion, R: Right testis volume, L: Left testis volume
In silico analysis
To further investigate the gene functions regulated by the differentially expressed miRs, target gene prediction was done using miRDB tool. GO and KEGG pathway analysis demonstrated the target genes of downregulated miRs with log2 fold change ≥2 to be enriched in important biological processes such as apoptosis and cell differentiation. Furthermore, the target genes were involved in important pathways such as phosphatidylinositol 3 kinase-protein kinase B (PI3K-Akt) and mitogen activated protein kinase (MAPK) signaling with significant enrichment scores as per the KEGG analysis [Figure 3].
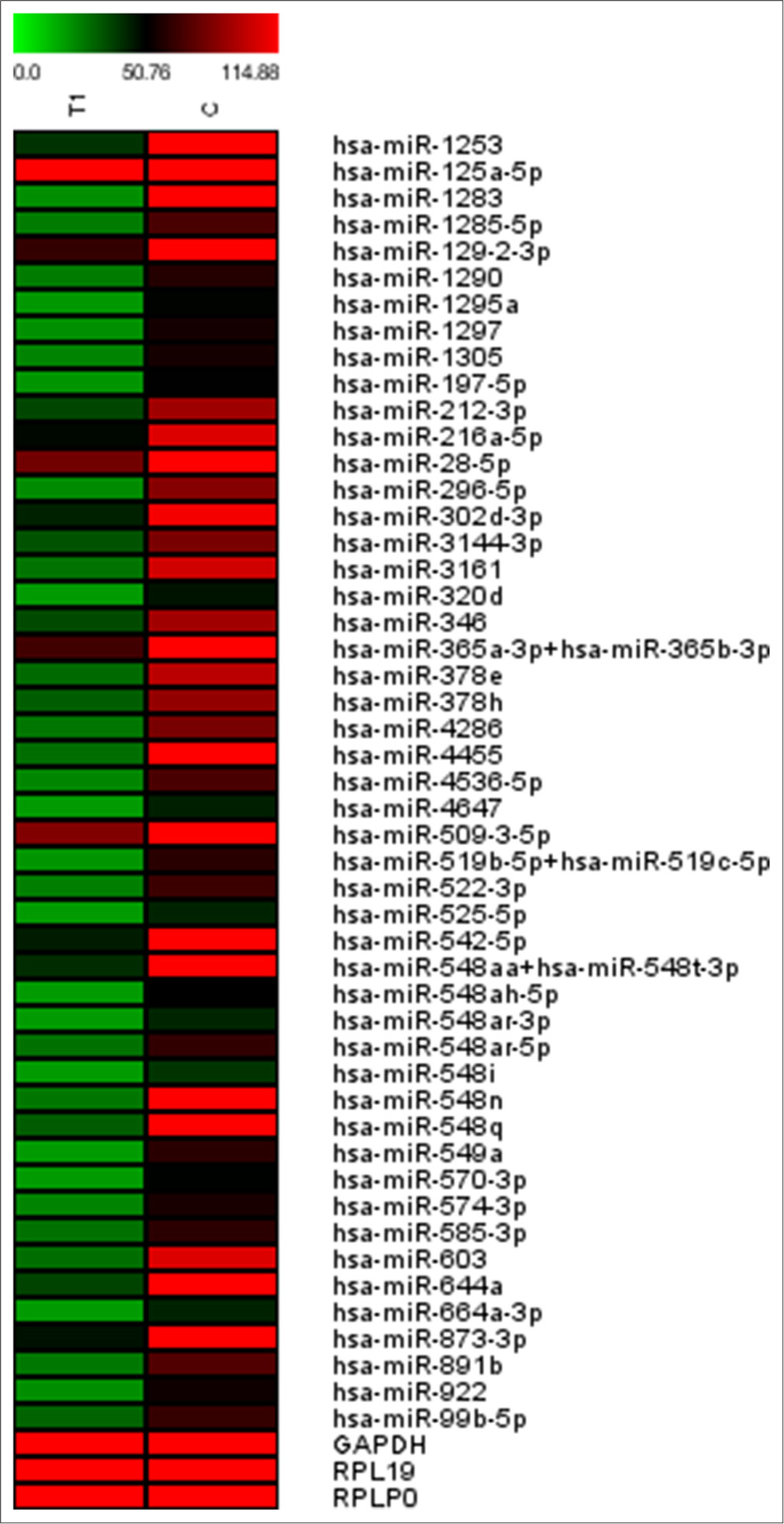
- Heatmap representation of DE miRs found in four iHS and four NS patients using nCounter miRNA expression analysis for validation. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase (for normalization control), RPL19: Ribosomal protein L19 (normalization control), RPLP0: Ribosomal protein lateral stalk subunit P0 (normalization control)
Validation
A distinct validation cohort of four iHS and four NS sample was used for nCounter miRNA expression analysis. We found miR-216a-3p, miR-522-3p, and miR-548aq as common DE miRs that are downregulated in iHS patients in experimental and validation cohorts [Figure 4]. The nCounter miRNA expression analysis showed 49 miRs showing DE, where three are found common in sequencing studies. The remaining 46 DE miRs profile need to be explored further and can be considered as DE miR profiles of testicular tissue in HS patients. The advantage of performing validation using a medium throughput technique like nCounter miRNA expression instead of quantitative polymerase chain reaction (qPCR) is acquiring a larger number of DE miRs along with validating the findings of high throughput techniques like sequencing. Although qPCR would have been a more specific technique of validation.
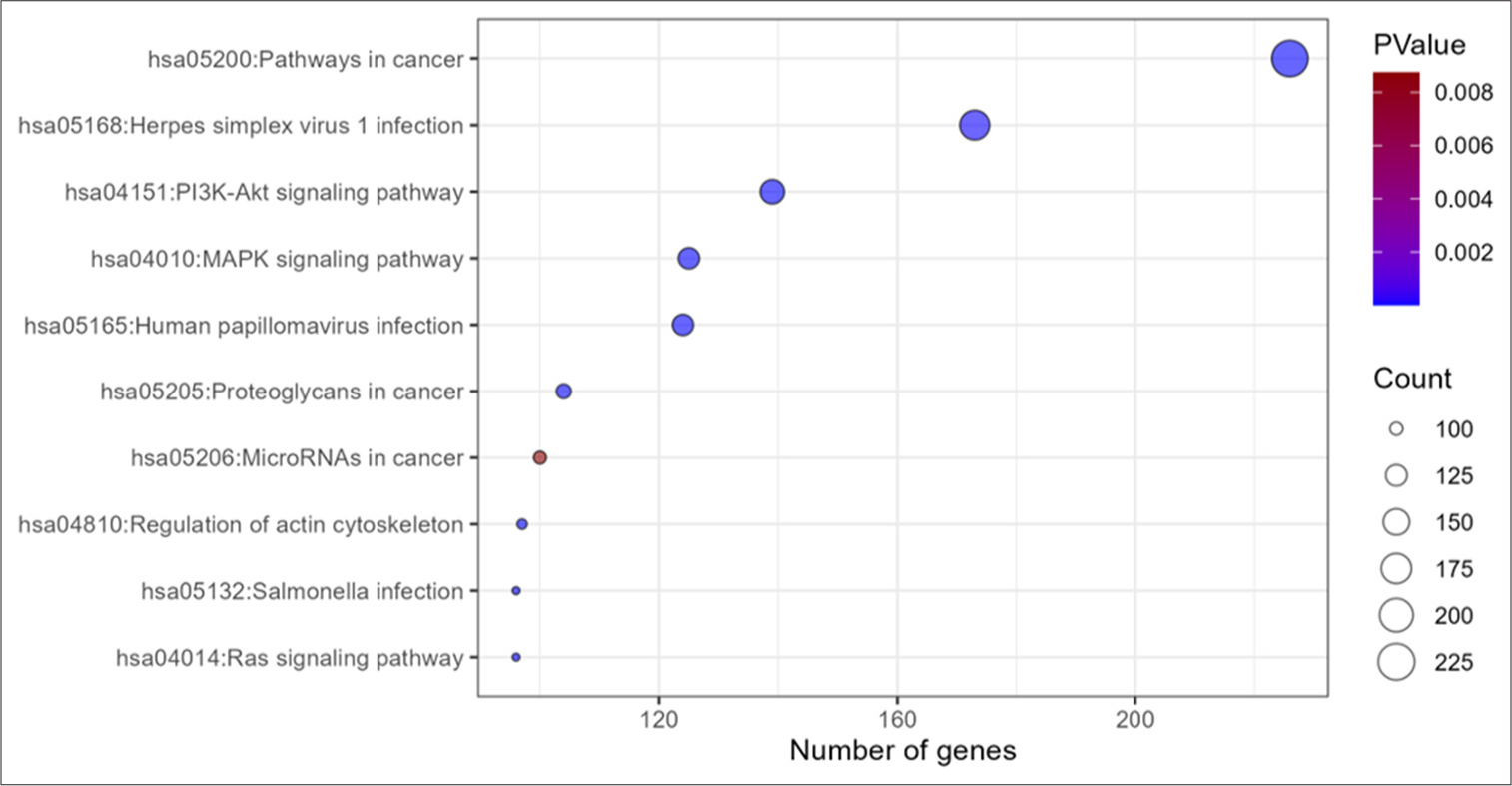
- KEGG pathway analysis from target gene list of miRs in iHS represented in bubble map. KEGG: Kyoto encyclopedia of genes and genomes, MAPK: Mitogen activated protein kinase, Pi3K-Akt: phosphatidylinositol 3-kinase protein kinase B, RNA: Ribonucleic acid
DISCUSSION
NOA affects 1% of men, with acquired, genetic, and epigenetic factors contributing to its clinical heterogeneity.[9,10] The spermatogenic failure, giving rise to an HS-like state of seminiferous epithelium, cannot be treated, and the patient must undergo assisted reproductive procedures. Sperm retrieval in HS patients gives up to a 90% success rate, which is the highest among all the subtypes of NOA.[11] Despite its prognostic importance, the existing literature lacks exploratory studies on epigenomic etiological factors associated with HS. In recent years, due to the advent of high-throughput sequencing technologies, the determination of molecular biomarkers and elucidation of molecular pathways implicated in idiopathic diseases has become possible. miR is a large family of small (approximately 21 nucleotides) non-coding RNAs that are involved in controlling the activity of around 30% of all protein-coding genes in almost every cellular process discovered so far.[12] Aberrant miR expression consistently correlated with infertility, suggesting their potential as biomarkers and for therapeutic development.[13] We performed small RNA sequencing-based profiling of miRs that are differentially expressed in HS. The DE miRs profile was compared with existing literature on SCOS and MA subtypes of NOA and identified dysregulation of five spermatogenic, which include one downregulated (miR-379-5p) and four upregulated (miR-449a, miR-181c, miR-34b-3p, and miR-122b-3p) miRNAs. In addition to a pooled analysis, we performed a comprehensive patient-wise evaluation of these five candidate spermatogenic miRs to assess the frequency of occurrence. We found eight (40%) of iHS patients to be associated with at least one of the candidate spermatogenic miRs. The upregulated miR, miR-379-5p, was observed in two patients. Downregulation of miR-34b-3p and miR-122b-5p was most prevalent and was observed in five HS patients, while that of miR-449a and miR-181c were observed in four patients. This finding shows the potential application of miRs in panel-based screening of NOA subtypes. The 12 iHS cases did not show any significant up- or down-regulation in candidate spermatogenic miRs, which suggests the involvement of unidentified mechanisms or potential candidate miRs with weak evidence for spermatogenesis regulation. We found one upregulated and 24 downregulated miRs in the non-overlapping profile of iHS patients, which needs to be validated via functional studies on cell line or animal models. The five candidate miRs, namely, miR-379-5p, miR-449a, miR-181c, miR-34b-3p, and miR-122b-3p, have been reported to perform a crucial role in the spermatogenetic pathway and the development of sperms. We assert the diagnostic importance and provide commentary drawing from relevant literature on these five spermatogenic miRs.
MiR-181
The miR-181 family comprises miR-181a, miR-181b, miR-181c, and miR-181d.[14] Among them, miR-181c is highly abundant in testis and spermatozoa, thus having a significant role in male fertility.[15] It regulates SMAD7 and RSBN1 expression and affects spermatogonia and spermatocytes differentiation.[16,17] Studies have shown its downregulation in HS and upregulation in conditions such as SCOS and AZFc deletion and higher expression correlates with successful sperm retrieval in NOA patients.[18] We found two HS samples showing downregulation of miR-181c with high and two with low fold change values. Moreover, the involvement of miR-181c in PI3K/Akt and Hippo pathways, both of which are crucial for spermatogenesis, suggests its role in azoospermia development.[19,20] The in silico analysis of our study showed apoptosis and cell differentiation to be the most enriched biological processes and the PI3K/Akt pathway to be significantly enriched when a downregulated miR expression profile was used for target prediction.
MiR-34/449
The miR-34 family, comprising miR-34a, miR-34b, and miR-34c, plays a significant role in spermatogenesis.[21] Studies show that miR-34c is involved in germ cell regulation, notably in spermatocytes and round spermatids, and its potential impact on apoptosis inhibition.[22,23] It may modulate pathways such as Notch, transforming growth factor beta (TGF-beta), and janus kinase 2 (JAK2)/Signal transducer and activator of transcription 3 (STAT3), which are critical for spermatogenesis. Dysregulation of miR-34c, observed in conditions like SCOS and MA, suggests its relevance in azoospermia pathogenesis.[24] We reported the association of miR-34c downregulation in HS patients with the highest occurrence among the identified candidate spermatogenic miRs. This indicates the diagnostic importance of miR-34c in NOA irrespective of a specific subtype.[8,25] The miR-449 is also a member of the miR-34/449 family, which is known to play a critical role in normal testicular function, spermatogenesis, and the regulation of sperm maturation and function. It is highly expressed in post-mitotic male germ cells. The deficiency of miR-449 disrupts both meiosis and the final stages of sperm maturation. Abnormal expression of miR-449 contributes to the development of conditions such as oligoasthenoteratozoospermia (a condition characterized by low sperm count, poor sperm motility, and abnormal sperm morphology) and infertility. Moreover, miR-449 has been associated with high sperm motility and normal sperm morphology when expressed at higher levels in sperm. Overexpression of miR-449, along with miR-26a, has been shown to inhibit sperm apoptosis and improve sperm motility.[26] Our findings strengthen the evidence for miR-34c and miR-449a to be associated with HS in the development of spermatogenic failure.
MiR-122
The miR-122 family of miRs also plays a role in male fertility and sperm quality regulation. The expression of miR-122-5p was lower in men with oligospermia/oligoasthenozoospermia and azoospermia compared to fertile men with normozoospermia.[27] This implies that miR-122-5p may have a role in spermatogenesis or sperm function, as its expression correlates with sperm concentration and motility.[28] Furthermore, the age-dependent decline in semen quality was associated with low expression of miR-122 in the seminal plasma of elderly men.
MiR-379
The miR-379-5p was observed to be upregulated in the HS patients of the present study. Previous research has demonstrated that miR-379-5p plays a crucial role in inhibiting the proliferation, invasion, and migration of tumor cells. It exerts its inhibitory effects by targeting various genes involved in tumor occurrence and development, such as insulin-like growth factor 1 (IGF-1), cyclin B1, and insulin like growth factor 1 receptor (IGF 1R).[29] Although it is reported to undergo significant downregulation in cancer tissues, including the liver, bladder, and breast cancer, its role in reproductive organ development has not been reported. However, given its known functions in inhibiting cell proliferation and its potential to regulate key pathways such as wingless-type mouse mammary tumor virus integration site (WNT), Hedgehog, and Hippo signaling pathways, it is speculated that miR-379-5p may also be involved in testis development and spermatogenesis.[30]
Availability of data and materials
The data presented in this study are available on reasonable request from the corresponding author
Limitation
The study is limited by technical constraints, specifically the insufficient availability of FNA samples for RNA isolation. Consequently, in this study, obtaining a sufficiently powered and well-matched control sample was expensive and time-consuming. The authors highly recommend the replication of this proof of concept study on a larger sample size. Therefore, the miR DE was assessed using three NS controls. All iHS patient samples were collected in a planned manner starting from the day when ethical approval was obtained, as there is no biobanking for storing testicular tissue samples available at the institute.
CONCLUSION
The DE of miRs emerges as a promising avenue for understanding the pathogenesis of HS. The objective was to identify candidate spermatogenic miRs associated with this condition. Our analysis identified dysregulated miRs in the spermatogenic family of miR-181c, miR-34/449 family, miR-122, and miR-379-5p that may serve as potential biomarkers for diagnostic screening. The identification of overlapping miRs between NOA and testicular HS indicates the common pathogenic origin of various histopathological subtypes. The spermatogenic failure of various extents found in HS, MA, and SCOS might represent the progressive nature of the disease sharing similar epigenetic profile of candidate spermatogenic miRs. The 55% of HS cases that remain idiopathic on miR profile analysis pose the need for more functional studies to identify novel spermatogenic miRs or epigenomic etiological factors associated with NOA. The in silico analysis-based molecular functions and signaling pathways provide insights into the molecular mechanisms underlying HS. Further research is warranted to validate these findings in larger cohorts and elucidate the functional significance of the identified epigenetic factors in HS pathophysiology.
Acknowledgment
The authors would like to sincerely thank the Indian Council of Medical Research for providing funding for this work. We acknowledge the effort of the technical staff of molecular cytogenetics staff at the Department of Reproductive Biology, All India Institute of Medical Sciences New Delhi, for providing support in performing XY-FISH and Yq microdeletion.
Authors’ contributions
NS: Investigation, formal analysis, writing – original draft; MJ: Conceptualization, methodology, resources, validation, writing – review and editing, supervision, funding acquisition; AH: Conceptualization, Supervisions; SK: Testicular FNA reposts; MK: Patient recruitment.
Ethical approval
The research/study approved by the Institutional Review Board at All India Institute of Medical Sciences in Delhi, number and date (IEC-15 1/06.03.2020, RP-17/2020).
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Conflicts of interest
Ashutosh Halder is on the Editorial Board of the Journal
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript, and no images were manipulated using AI.
Supplementary data available on:
Financial support and sponsorship: The work is funded by the Indian Council of Medical Research (ICMR), India.
References
- Worldwide trend analysis of primary and secondary infertility rates over past decades: A cross-sectional study. Int J Reprod Biomed. 2022;20:37-46.
- [CrossRef] [PubMed] [Google Scholar]
- A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13:37.
- [CrossRef] [PubMed] [Google Scholar]
- Histopathological patterns of testicular biopsy in male infertility: A retrospective study from a tertiary care center in the western part of Saudi Arabia. Urol Ann. 2011;3:19-23.
- [CrossRef] [PubMed] [Google Scholar]
- Testicular histology may predict the successful sperm retrieval in patients with non-obstructive azoospermia undergoing conventional TESE: A diagnostic accuracy study. J Assisted Reprod Genet. 2017;34:149-54.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA expression profiles in testicular biopsies of patients with impaired spermatogenesis. Andrology. 2016;4:1020-7.
- [CrossRef] [PubMed] [Google Scholar]
- Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol. 2009;7:13.
- [CrossRef] [PubMed] [Google Scholar]
- Altered microRNA profiles of testicular biopsies from patients with nonobstructive azoospermia. Asian J Androl. 2020;22:100-5.
- [CrossRef] [PubMed] [Google Scholar]
- Panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility. Fertil Steril. 2014;102:989-97.e1.
- [CrossRef] [PubMed] [Google Scholar]
- microRNAs in the pathogenesis of non-obstructive azoospermia: The underlying mechanisms and therapeutic potentials. Syst Biol Reprod Med. 2021;67:337-53.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNAs in spermatogenesis dysfunction and male infertility: Clinical phenotypes, mechanisms and potential diagnostic biomarkers. Front Endocrinol. 2024;15:1293368.
- [CrossRef] [PubMed] [Google Scholar]
- Outcomes and predictive factors of successful salvage microdissection testicular sperm extraction (mTESE) after failed classic TESE: Results from a multicenter cross-sectional study. Int J Impotence Res. 2022;34:795-9.
- [CrossRef] [PubMed] [Google Scholar]
- The role of miRNAs in male human reproduction: A systematic review. Andrology. 2020;8:7-26.
- [CrossRef] [PubMed] [Google Scholar]
- Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 2013;41:4104-17.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer. 2014;13:86.
- [CrossRef] [PubMed] [Google Scholar]
- SMAD expression in the testis: An insight into BMP regulation of spermatogenesis. Dev Dyn. 2008;237:97-111.
- [CrossRef] [PubMed] [Google Scholar]
- Role of microRNAs in mammalian spermatogenesis and testicular germ cell tumors. Reproduction. 2015;149:R127-37.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA profile comparison of testicular tissues derived from successful and unsuccessful microdissection testicular sperm extraction retrieval in non-obstructive azoospermia patients. Reprod Fertil Dev. 2019;31:671-82.
- [CrossRef] [PubMed] [Google Scholar]
- Spermatogenesis affects the outcome of ICSI for azoospermic patients rather than sperm retrieval method. Syst Biol Reprod Med. 2010;56:457-64.
- [CrossRef] [PubMed] [Google Scholar]
- LncRNA SNHG6 functions as a ceRNA to regulate neuronal cell apoptosis by modulating miR-181c-5p/BIM signalling in ischaemic stroke. J Cell Mol Med. 2022;23:6120-30.
- [CrossRef] [PubMed] [Google Scholar]
- Expression of miR-34c induces G2/M cell cycle arrest in breast cancer cells. BMC Cancer. 2014;14:538.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS One. 2012;7:e33861.
- [CrossRef] [PubMed] [Google Scholar]
- Oligoasthenoteratozoospermia and infertility in mice deficient for miR-34b/c and miR-449 loci. PLoS Genet. 2014;10:e1004597.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of deleterious NOTCH mutation as novel predictor to efficacious immunotherapy in NSCLC. Clin Cancer Res. 2020;26:3649-61.
- [CrossRef] [PubMed] [Google Scholar]
- MiR-410 affects the proliferation and apoptosis of lung cancer A549 cells through regulation of SOCS3/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24:11462.
- [Google Scholar]
- Association between sperm morphology and altered sperm microRNA expression. Biology. 2022;11:1671.
- [CrossRef] [PubMed] [Google Scholar]
- Qualitative and quantitative assessment of sperm miRNAs identifies hsa-miR-9-3p, hsa-miR-30b-5p and hsamiR-122-5p as potential biomarkers of male infertility and sperm quality. Reprod Biol Endocrinol. 2022;20:122.
- [CrossRef] [PubMed] [Google Scholar]
- MiR-125b-2 knockout in testis is associated with targeting to the PAP gene, mitochondrial copy number, and impaired sperm quality. Int J Mol Sci. 2019;20:148.
- [CrossRef] [PubMed] [Google Scholar]
- miR-379 regulates cyclin B1 expression and is decreased in breast cancer. PLoS One. 2013;8:e68753.
- [CrossRef] [PubMed] [Google Scholar]
- Dynamic Hedgehog signalling pathway activity in germline stem cells. Andrology. 2014;2:267-74.
- [CrossRef] [PubMed] [Google Scholar]







